ClearSee: a rapid optical clearing reagent for whole-plant fluorescence imaging
- PMID: 26493404
- PMCID: PMC4712841
- DOI: 10.1242/dev.127613
ClearSee: a rapid optical clearing reagent for whole-plant fluorescence imaging
Abstract
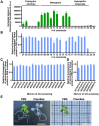
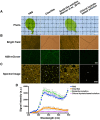
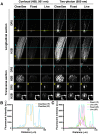
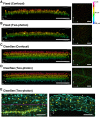
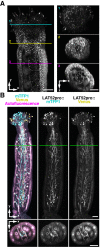
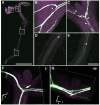
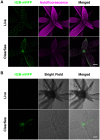
Imaging techniques for visualizing and analyzing precise morphology and gene expression patterns are essential for understanding biological processes during development in all organisms. With the aid of chemical screening, we developed a clearing method using chemical solutions, termed ClearSee, for deep imaging of morphology and gene expression in plant tissues. ClearSee rapidly diminishes chlorophyll autofluorescence while maintaining fluorescent protein stability. By adjusting the refractive index mismatch, whole-organ and whole-plant imaging can be performed by both confocal and two-photon excitation microscopy in ClearSee-treated samples. Moreover, ClearSee is applicable to multicolor imaging of fluorescent proteins to allow structural analysis of multiple gene expression. Given that ClearSee is compatible with staining by chemical dyes, the technique is useful for deep imaging in conjunction with genetic markers and for plant species not amenable to transgenic approaches. This method is useful for whole imaging for intact morphology and will help to accelerate the discovery of new phenomena in plant biological research.
Keywords: Arabidopsis thaliana; Clearing; Confocal microscopy; Deep imaging; Physcomitrella patens; Two-photon microscopy; Whole plant.
© 2015. Published by The Company of Biologists Ltd.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures

Similar articles
-
A protocol for combining fluorescent proteins with histological stains for diverse cell wall components.Plant J. 2018 Jan;93(2):399-412. doi: 10.1111/tpj.13784. Plant J. 2018. PMID: 29171896
-
ClearSeeAlpha: Advanced Optical Clearing for Whole-Plant Imaging.Plant Cell Physiol. 2021 Nov 10;62(8):1302-1310. doi: 10.1093/pcp/pcab033. Plant Cell Physiol. 2021. PMID: 33638989 Free PMC article.
-
Optical Clearing of Plant Tissues for Fluorescence Imaging.J Vis Exp. 2022 Jan 5;(179). doi: 10.3791/63428. J Vis Exp. 2022. PMID: 35068484
-
Advances in Two-Photon Imaging in Plants.Plant Cell Physiol. 2021 Nov 10;62(8):1224-1230. doi: 10.1093/pcp/pcab062. Plant Cell Physiol. 2021. PMID: 34019083 Free PMC article. Review.
-
Light microscopy of whole plant organs.J Microsc. 2016 Aug;263(2):165-70. doi: 10.1111/jmi.12394. Epub 2016 Mar 29. J Microsc. 2016. PMID: 27027806 Review.
Cited by
-
Tissue growth constrains root organ outlines into an isometrically scalable shape.Development. 2021 Feb 26;148(4):dev196253. doi: 10.1242/dev.196253. Development. 2021. PMID: 33637613 Free PMC article.
-
The diversity of stomatal development regulation in Callitriche is related to the intrageneric diversity in lifestyles.Proc Natl Acad Sci U S A. 2021 Apr 6;118(14):e2026351118. doi: 10.1073/pnas.2026351118. Proc Natl Acad Sci U S A. 2021. PMID: 33782136 Free PMC article.
-
A proxitome-RNA-capture approach reveals that processing bodies repress coregulated hub genes.Plant Cell. 2024 Feb 26;36(3):559-584. doi: 10.1093/plcell/koad288. Plant Cell. 2024. PMID: 37971938 Free PMC article.
-
An Improved Recombineering Toolset for Plants.Plant Cell. 2020 Jan;32(1):100-122. doi: 10.1105/tpc.19.00431. Epub 2019 Oct 30. Plant Cell. 2020. PMID: 31666295 Free PMC article.
-
Spatiotemporal plant hormone analysis from cryosections using laser microdissection-liquid chromatography-mass spectrometry.J Plant Res. 2022 Mar;135(2):377-386. doi: 10.1007/s10265-021-01360-x. Epub 2021 Nov 23. J Plant Res. 2022. PMID: 34812978
References
-
- Adachi S., Minamisawa K., Okushima Y., Inagaki S., Yoshiyama K., Kondou Y., Kaminuma E., Kawashima M., Toyoda T., Matsui M. et al. (2011). Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proc. Natl. Acad. Sci. USA 108, 10004-10009. 10.1073/pnas.1103584108 - DOI - PMC - PubMed
-
- Bilsborough G. D., Runions A., Barkoulas M., Jenkins H. W., Hasson A., Galinha C., Laufs P., Hay A., Prusinkiewicz P. and Tsiantis M. (2011). Model for the regulation of Arabidopsis thaliana leaf margin development. Proc. Natl. Acad. Sci. USA 108, 3424-3429. 10.1073/pnas.1015162108 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

