PDB-Explorer: a web-based interactive map of the protein data bank in shape space
- PMID: 26493835
- PMCID: PMC4619230
- DOI: 10.1186/s12859-015-0776-9
PDB-Explorer: a web-based interactive map of the protein data bank in shape space
Abstract
Background: The RCSB Protein Data Bank (PDB) provides public access to experimentally determined 3D-structures of biological macromolecules (proteins, peptides and nucleic acids). While various tools are available to explore the PDB, options to access the global structural diversity of the entire PDB and to perceive relationships between PDB structures remain very limited.
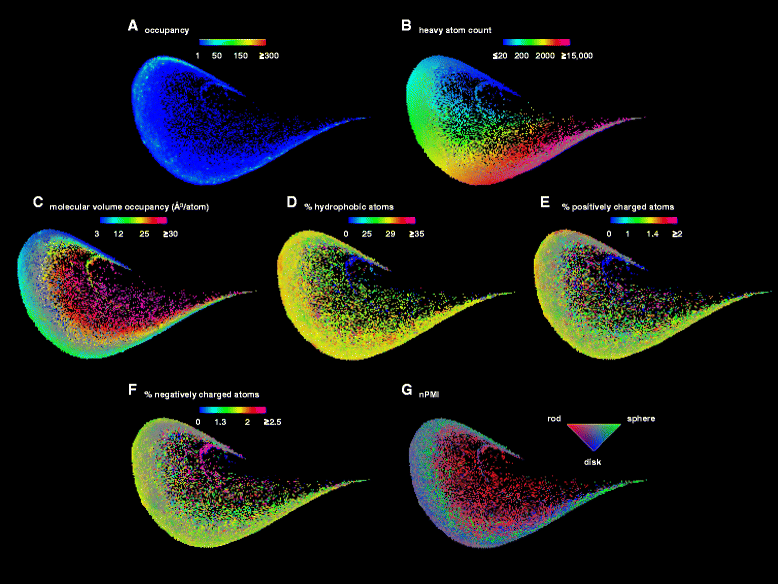
Methods: A 136-dimensional atom pair 3D-fingerprint for proteins (3DP) counting categorized atom pairs at increasing through-space distances was designed to represent the molecular shape of PDB-entries. Nearest neighbor searches examples were reported exemplifying the ability of 3DP-similarity to identify closely related biomolecules from small peptides to enzyme and large multiprotein complexes such as virus particles. The principle component analysis was used to obtain the visualization of PDB in 3DP-space.
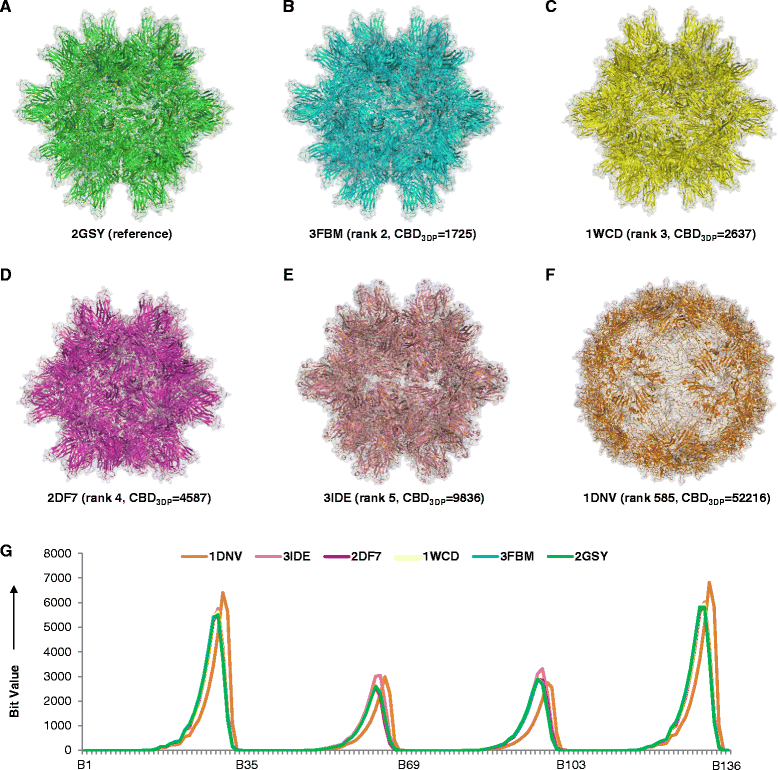
Results: The 3DP property space groups proteins and protein assemblies according to their 3D-shape similarity, yet shows exquisite ability to distinguish between closely related structures. An interactive website called PDB-Explorer is presented featuring a color-coded interactive map of PDB in 3DP-space. Each pixel of the map contains one or more PDB-entries which are directly visualized as ribbon diagrams when the pixel is selected. The PDB-Explorer website allows performing 3DP-nearest neighbor searches of any PDB-entry or of any structure uploaded as protein-type PDB file. All functionalities on the website are implemented in JavaScript in a platform-independent manner and draw data from a server that is updated daily with the latest PDB additions, ensuring complete and up-to-date coverage. The essentially instantaneous 3DP-similarity search with the PDB-Explorer provides results comparable to those of much slower 3D-alignment algorithms, and automatically clusters proteins from the same superfamilies in tight groups.
Conclusion: A chemical space classification of PDB based on molecular shape was obtained using a new atom-pair 3D-fingerprint for proteins and implemented in a web-based database exploration tool comprising an interactive color-coded map of the PDB chemical space and a nearest neighbor search tool. The PDB-Explorer website is freely available at www.cheminfo.org/pdbexplorer and represents an unprecedented opportunity to interactively visualize and explore the structural diversity of the PDB. ᅟ
Figures









References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

