MicroRNA-encoded behavior in Drosophila
- PMID: 26494171
- PMCID: PMC4902127
- DOI: 10.1126/science.aad0217
MicroRNA-encoded behavior in Drosophila
Abstract
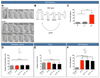
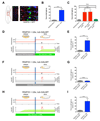
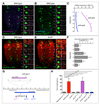
The relationship between microRNA (miRNA) regulation and the specification of behavior is only beginning to be explored. We found that mutation of a single miRNA locus (miR-iab4/iab8) in Drosophila larvae affects the animal's capacity to correct its orientation if turned upside down (self-righting). One of the miRNA targets involved in this behavior is the Hox gene Ultrabithorax, whose derepression in two metameric neurons leads to self-righting defects. In vivo neural activity analysis reveals that these neurons, the self-righting node (SRN), have different activity patterns in wild type and miRNA mutants, whereas thermogenetic manipulation of SRN activity results in changes in self-righting behavior. Our work thus reveals a miRNA-encoded behavior and suggests that other miRNAs might also be involved in behavioral control in Drosophila and other species.
Copyright © 2015, American Association for the Advancement of Science.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

