Overshoot during phenotypic switching of cancer cell populations
- PMID: 26494317
- PMCID: PMC4616026
- DOI: 10.1038/srep15464
Overshoot during phenotypic switching of cancer cell populations
Abstract
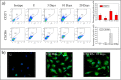
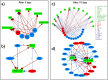
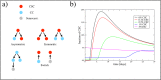
The dynamics of tumor cell populations is hotly debated: do populations derive hierarchically from a subpopulation of cancer stem cells (CSCs), or are stochastic transitions that mutate differentiated cancer cells to CSCs important? Here we argue that regulation must also be important. We sort human melanoma cells using three distinct cancer stem cell (CSC) markers - CXCR6, CD271 and ABCG2 - and observe that the fraction of non-CSC-marked cells first overshoots to a higher level and then returns to the level of unsorted cells. This clearly indicates that the CSC population is homeostatically regulated. Combining experimental measurements with theoretical modeling and numerical simulations, we show that the population dynamics of cancer cells is associated with a complex miRNA network regulating the Wnt and PI3K pathways. Hence phenotypic switching is not stochastic, but is tightly regulated by the balance between positive and negative cells in the population. Reducing the fraction of CSCs below a threshold triggers massive phenotypic switching, suggesting that a therapeutic strategy based on CSC eradication is unlikely to succeed.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
-
- Bonnet D. & Dick J. E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 3, 730–737 (1997). - PubMed
-
- La Porta C. A. M. & Zapperi S. Human breast and melanoma cancer stem cells biomarkers. Cancer Lett 338, 69–73 (2013). - PubMed
-
- Schepers A. G. et al. Lineage tracing reveals lgr5+ stem cell activity in mouse intestinal adenomas. Science 337, 730–5 (2012). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

