JMJD1C is required for the survival of acute myeloid leukemia by functioning as a coactivator for key transcription factors
- PMID: 26494788
- PMCID: PMC4617977
- DOI: 10.1101/gad.267278.115
JMJD1C is required for the survival of acute myeloid leukemia by functioning as a coactivator for key transcription factors
Abstract
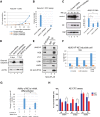
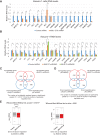
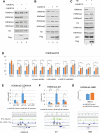
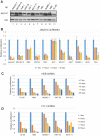
RUNX1-RUNX1T1 (formerly AML1-ETO), a transcription factor generated by the t(8;21) translocation in acute myeloid leukemia (AML), dictates a leukemic program by increasing self-renewal and inhibiting differentiation. Here we demonstrate that the histone demethylase JMJD1C functions as a coactivator for RUNX1-RUNX1T1 and is required for its transcriptional program. JMJD1C is directly recruited by RUNX1-RUNX1T1 to its target genes and regulates their expression by maintaining low H3K9 dimethyl (H3K9me2) levels. Analyses in JMJD1C knockout mice also establish a JMJD1C requirement for RUNX1-RUNX1T1's ability to increase proliferation. We also show a critical role for JMJD1C in the survival of multiple human AML cell lines, suggesting that it is required for leukemic programs in different AML cell types through its association with key transcription factors.
Keywords: AML1-ETO; HEB; LYL1; acute myeloid leukemia; histone demethylase; transcription regulation.
© 2015 Chen et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



Similar articles
-
Critical role of Jumonji domain of JMJD1C in MLL-rearranged leukemia.Blood Adv. 2019 May 14;3(9):1499-1511. doi: 10.1182/bloodadvances.2018026054. Blood Adv. 2019. PMID: 31076406 Free PMC article.
-
JMJD1C forms condensate to facilitate a RUNX1-dependent gene expression program shared by multiple types of AML cells.Protein Cell. 2025 May 28;16(5):338-364. doi: 10.1093/procel/pwae059. Protein Cell. 2025. PMID: 39450904 Free PMC article.
-
JMJD1C-mediated metabolic dysregulation contributes to HOXA9-dependent leukemogenesis.Leukemia. 2019 Jun;33(6):1400-1410. doi: 10.1038/s41375-018-0354-z. Epub 2019 Jan 8. Leukemia. 2019. PMID: 30622285
-
RUNX1-ETO Leukemia.Adv Exp Med Biol. 2017;962:151-173. doi: 10.1007/978-981-10-3233-2_11. Adv Exp Med Biol. 2017. PMID: 28299657 Review.
-
Molecular pathways mediating MDS/AML with focus on AML1/RUNX1 point mutations.J Cell Physiol. 2009 Jul;220(1):16-20. doi: 10.1002/jcp.21769. J Cell Physiol. 2009. PMID: 19334039 Review.
Cited by
-
Critical role of Jumonji domain of JMJD1C in MLL-rearranged leukemia.Blood Adv. 2019 May 14;3(9):1499-1511. doi: 10.1182/bloodadvances.2018026054. Blood Adv. 2019. PMID: 31076406 Free PMC article.
-
Biology and targeting of the Jumonji-domain histone demethylase family in childhood neoplasia: a preclinical overview.Expert Opin Ther Targets. 2019 Apr;23(4):267-280. doi: 10.1080/14728222.2019.1580692. Epub 2019 Feb 26. Expert Opin Ther Targets. 2019. PMID: 30759030 Free PMC article. Review.
-
New insights into transcriptional and leukemogenic mechanisms of AML1-ETO and E2A fusion proteins.Front Biol (Beijing). 2016 Aug;11(4):285-304. doi: 10.1007/s11515-016-1415-1. Epub 2016 Sep 3. Front Biol (Beijing). 2016. PMID: 28261265 Free PMC article.
-
Crucial Functions of the JMJD1/KDM3 Epigenetic Regulators in Cancer.Mol Cancer Res. 2021 Jan;19(1):3-13. doi: 10.1158/1541-7786.MCR-20-0404. Epub 2020 Jun 30. Mol Cancer Res. 2021. PMID: 32605929 Free PMC article. Review.
-
Development of an in vitro genotoxicity assay to detect retroviral vector-induced lymphoid insertional mutants.Mol Ther Methods Clin Dev. 2023 Aug 22;30:515-533. doi: 10.1016/j.omtm.2023.08.017. eCollection 2023 Sep 14. Mol Ther Methods Clin Dev. 2023. PMID: 37693949 Free PMC article.
References
-
- Ben-Ami O, Friedman D, Leshkowitz D, Goldenberg D, Orlovsky K, Pencovich N, Lotem J, Tanay A, Groner Y. 2013. Addiction of t(8;21) and inv(16) acute myeloid leukemia to native RUNX1. Cell Rep 4: 1131–1143. - PubMed
-
- Cabezas-Wallscheid N, Eichwald V, de Graaf J, Lower M, Lehr HA, Kreft A, Eshkind L, Hildebrandt A, Abassi Y, Heck R, et al. 2013. Instruction of haematopoietic lineage choices, evolution of transcriptional landscapes and cancer stem cell hierarchies derived from an AML1-ETO mouse model. EMBO Mol Med 5: 1804–1820. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous
