An Exploration of the Universe of Polyglutamine Structures
- PMID: 26495838
- PMCID: PMC4619799
- DOI: 10.1371/journal.pcbi.1004541
An Exploration of the Universe of Polyglutamine Structures
Abstract
Deposits of misfolded proteins in the human brain are associated with the development of many neurodegenerative diseases. Recent studies show that these proteins have common traits even at the monomer level. Among them, a polyglutamine region that is present in huntingtin is known to exhibit a correlation between the length of the chain and the severity as well as the earliness of the onset of Huntington disease. Here, we apply bias exchange molecular dynamics to generate structures of polyglutamine expansions of several lengths and characterize the resulting independent conformations. We compare the properties of these conformations to those of the standard proteins, as well as to other homopolymeric tracts. We find that, similar to the previously studied polyvaline chains, the set of possible transient folds is much broader than the set of known-to-date folds, although the conformations have different structures. We show that the mechanical stability is not related to any simple geometrical characteristics of the structures. We demonstrate that long polyglutamine expansions result in higher mechanical stability than the shorter ones. They also have a longer life span and are substantially more prone to form knotted structures. The knotted region has an average length of 35 residues, similar to the typical threshold for most polyglutamine-related diseases. Similarly, changes in shape and mechanical stability appear once the total length of the peptide exceeds this threshold of 35 glutamine residues. We suggest that knotted conformers may also harm the cellular machinery and thus lead to disease.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
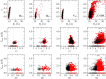
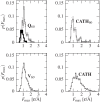
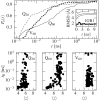
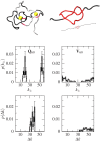
Similar articles
-
Effects of chain length on the aggregation of model polyglutamine peptides: molecular dynamics simulations.Proteins. 2007 Jan 1;66(1):96-109. doi: 10.1002/prot.21132. Proteins. 2007. PMID: 17068817
-
Polyglutamine induced misfolding of huntingtin exon1 is modulated by the flanking sequences.PLoS Comput Biol. 2010 Apr 29;6(4):e1000772. doi: 10.1371/journal.pcbi.1000772. PLoS Comput Biol. 2010. PMID: 20442863 Free PMC article.
-
Transformation between α-helix and β-sheet structures of one and two polyglutamine peptides in explicit water molecules by replica-exchange molecular dynamics simulations.J Comput Chem. 2014 Jul 15;35(19):1430-7. doi: 10.1002/jcc.23633. Epub 2014 May 16. J Comput Chem. 2014. PMID: 24831733
-
Transient knots in intrinsically disordered proteins and neurodegeneration.Prog Mol Biol Transl Sci. 2020;174:79-103. doi: 10.1016/bs.pmbts.2020.03.003. Epub 2020 Apr 9. Prog Mol Biol Transl Sci. 2020. PMID: 32828471 Review.
-
Properties of polyglutamine expansion in vitro and in a cellular model for Huntington's disease.Philos Trans R Soc Lond B Biol Sci. 1999 Jun 29;354(1386):1013-9. doi: 10.1098/rstb.1999.0453. Philos Trans R Soc Lond B Biol Sci. 1999. PMID: 10434300 Free PMC article. Review.
Cited by
-
Conformational entropy limits the transition from nucleation to elongation in amyloid aggregation.Biophys J. 2022 Aug 2;121(15):2931-2939. doi: 10.1016/j.bpj.2022.06.031. Epub 2022 Jul 1. Biophys J. 2022. PMID: 35778843 Free PMC article.
-
Topological transformations in proteins: effects of heating and proximity of an interface.Sci Rep. 2017 Jan 4;7:39851. doi: 10.1038/srep39851. Sci Rep. 2017. PMID: 28051124 Free PMC article.
-
Unfolding of α-helical 20-residue poly-glutamic acid analyzed by multiple runs of canonical molecular dynamics simulations.PeerJ. 2018 May 15;6:e4769. doi: 10.7717/peerj.4769. eCollection 2018. PeerJ. 2018. PMID: 29780670 Free PMC article.
-
Effects of the enlargement of polyglutamine segments on the structure and folding of ataxin-2 and ataxin-3 proteins.J Biomol Struct Dyn. 2017 Feb;35(3):504-519. doi: 10.1080/07391102.2016.1152199. Epub 2016 May 20. J Biomol Struct Dyn. 2017. PMID: 26861241 Free PMC article.
References
-
- Chothia C, Finkelstein AV. The classification and origins of protein folding patterns. Annual Review of Biochemistry. 1990;59(1):1007–1035. - PubMed
-
- Sillitoe I, Cuff AL, Dessailly BH, Dawson NL, Furnham N, Lee D, et al. New functional families (FunFams) in CATH to improve the mapping of conserved functional sites to 3D structures. Nucleic Acids Research. 2013;41(D1):D490–D498. Available from: http://nar.oxfordjournals.org/content/41/D1/D490.abstract. 10.1093/nar/gks1211 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

