Mechanisms of drug resistance: daptomycin resistance
- PMID: 26495887
- PMCID: PMC4966536
- DOI: 10.1111/nyas.12948
Mechanisms of drug resistance: daptomycin resistance
Abstract
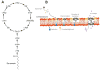
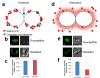
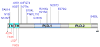
Daptomycin (DAP) is a cyclic lipopeptide with in vitro activity against a variety of Gram-positive pathogens, including multidrug-resistant organisms. Since its introduction into clinical practice in 2003, DAP has become an important key frontline antibiotic for severe or deep-seated infections caused by Gram-positive organisms. Unfortunately, DAP resistance (DAP-R) has been extensively documented in clinically important organisms such as Staphylococcus aureus, Enterococcus spp., and Streptococcus spp. Studies on the mechanisms of DAP-R in Bacillus subtilis and other Gram-positive bacteria indicate that the genetic pathways of DAP-R are diverse and complex. However, a common phenomenon emerging from these mechanistic studies is that DAP-R is associated with important adaptive changes in cell wall and cell membrane homeostasis with critical changes in cell physiology. Findings related to these adaptive changes have provided novel insights into the genetics and molecular mechanisms of bacterial cell envelope stress response and the manner in which Gram-positive bacteria cope with the antimicrobial peptide attack and protect vital structures of the cell envelope, such as the cell membrane. In this review, we will examine the most recent findings related to the molecular mechanisms of resistance to DAP in relevant Gram-positive pathogens and discuss the clinical implications for therapy against these important bacteria.
Keywords: Bacillus subtilis; Enterococcus; Staphylococcus aureus; daptomycin resistance.
© 2015 New York Academy of Sciences.
Figures


Similar articles
-
Daptomycin-resistant Enterococcus faecalis diverts the antibiotic molecule from the division septum and remodels cell membrane phospholipids.mBio. 2013 Jul 23;4(4):e00281-13. doi: 10.1128/mBio.00281-13. mBio. 2013. PMID: 23882013 Free PMC article.
-
Whole-genome analysis of a daptomycin-susceptible enterococcus faecium strain and its daptomycin-resistant variant arising during therapy.Antimicrob Agents Chemother. 2013 Jan;57(1):261-8. doi: 10.1128/AAC.01454-12. Epub 2012 Oct 31. Antimicrob Agents Chemother. 2013. PMID: 23114757 Free PMC article.
-
Antibacterial mechanism of daptomycin antibiotic against Staphylococcus aureus based on a quantitative bacterial proteome analysis.J Proteomics. 2017 Jan 6;150:242-251. doi: 10.1016/j.jprot.2016.09.014. Epub 2016 Sep 29. J Proteomics. 2017. PMID: 27693894
-
The efficacy and safety of daptomycin: first in a new class of antibiotics for Gram-positive bacteria.Clin Microbiol Infect. 2006 Mar;12 Suppl 1:24-32. doi: 10.1111/j.1469-0691.2006.01342.x. Clin Microbiol Infect. 2006. PMID: 16445721 Review.
-
Mechanism of Action and Resistance to Daptomycin in Staphylococcus aureus and Enterococci.Cold Spring Harb Perspect Med. 2016 Nov 1;6(11):a026997. doi: 10.1101/cshperspect.a026997. Cold Spring Harb Perspect Med. 2016. PMID: 27580748 Free PMC article. Review.
Cited by
-
Complete Genome Sequence of Enterococcus hirae R17, a Daptomycin-Resistant Bacterium Isolated from Retail Pork in China.Genome Announc. 2016 Jun 23;4(3):e00605-16. doi: 10.1128/genomeA.00605-16. Genome Announc. 2016. PMID: 27340071 Free PMC article.
-
Daptomycin: an evidence-based review of its role in the treatment of Gram-positive infections.Infect Drug Resist. 2016 Apr 15;9:47-58. doi: 10.2147/IDR.S99046. eCollection 2016. Infect Drug Resist. 2016. PMID: 27143941 Free PMC article. Review.
-
Polymyxin and lipopeptide antibiotics: membrane-targeting drugs of last resort.Microbiology (Reading). 2022 Feb;168(2):001136. doi: 10.1099/mic.0.001136. Microbiology (Reading). 2022. PMID: 35118938 Free PMC article. Review.
-
Staph wars: the antibiotic pipeline strikes back.Microbiology (Reading). 2023 Sep;169(9):001387. doi: 10.1099/mic.0.001387. Microbiology (Reading). 2023. PMID: 37656158 Free PMC article. Review.
-
Clinical Pharmacokinetics of Daptomycin.Clin Pharmacokinet. 2021 Mar;60(3):271-281. doi: 10.1007/s40262-020-00968-x. Epub 2020 Dec 14. Clin Pharmacokinet. 2021. PMID: 33313994 Review.
References
-
- Review on Antimicrobial Resistance. Antimicrobial resistance: Tackling a crisis for the future health and wealth of nations. [March 11 2015];2014 http://amr-review.org/
-
- World Health Organization. Antimicrobial resistance: global report on surveillance 2014. [March 4 2015];2014 http://www.who.int/drugresistance/documents/surveillancereport/en/
-
- Centers for Disease Control and Prevention. Antibiotic resistance threats in the United States. [March 9 2015];2013 http://www.cdc.gov/drugresistance/threat-report2013/index.html.
-
- Cosgrove SE, Corey GR. A balancing act: microbe versus muscle. Clin Infect Dis. 2009;49:181–183. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

