Identification of a Novel Lipoprotein Regulator of Clostridium difficile Spore Germination
- PMID: 26496694
- PMCID: PMC4619724
- DOI: 10.1371/journal.ppat.1005239
Identification of a Novel Lipoprotein Regulator of Clostridium difficile Spore Germination
Abstract
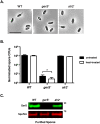
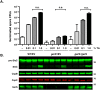
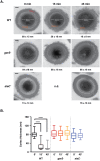
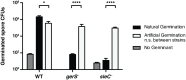
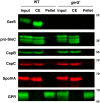
Clostridium difficile is a Gram-positive spore-forming pathogen and a leading cause of nosocomial diarrhea. C. difficile infections are transmitted when ingested spores germinate in the gastrointestinal tract and transform into vegetative cells. Germination begins when the germinant receptor CspC detects bile salts in the gut. CspC is a subtilisin-like serine pseudoprotease that activates the related CspB serine protease through an unknown mechanism. Activated CspB cleaves the pro-SleC zymogen, which allows the activated SleC cortex hydrolase to degrade the protective cortex layer. While these regulators are essential for C. difficile spores to outgrow and form toxin-secreting vegetative cells, the mechanisms controlling their function have only been partially characterized. In this study, we identify the lipoprotein GerS as a novel regulator of C. difficile spore germination using targeted mutagenesis. A gerS mutant has a severe germination defect and fails to degrade cortex even though it processes SleC at wildtype levels. Using complementation analyses, we demonstrate that GerS secretion, but not lipidation, is necessary for GerS to activate SleC. Importantly, loss of GerS attenuates the virulence of C. difficile in a hamster model of infection. Since GerS appears to be conserved exclusively in related Peptostreptococcaeace family members, our results contribute to a growing body of work indicating that C. difficile has evolved distinct mechanisms for controlling the exit from dormancy relative to B. subtilis and other spore-forming organisms.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Clostridium difficile Lipoprotein GerS Is Required for Cortex Modification and Thus Spore Germination.mSphere. 2018 Jun 27;3(3):e00205-18. doi: 10.1128/mSphere.00205-18. Print 2018 Jun 27. mSphere. 2018. PMID: 29950380 Free PMC article.
-
Revisiting the Role of Csp Family Proteins in Regulating Clostridium difficile Spore Germination.J Bacteriol. 2017 Oct 17;199(22):e00266-17. doi: 10.1128/JB.00266-17. Print 2017 Nov 15. J Bacteriol. 2017. PMID: 28874406 Free PMC article.
-
Regulation of Clostridium difficile spore germination by the CspA pseudoprotease domain.Biochimie. 2016 Mar;122:243-54. doi: 10.1016/j.biochi.2015.07.023. Epub 2015 Jul 29. Biochimie. 2016. PMID: 26231446 Free PMC article.
-
A Revised Understanding of Clostridioides difficile Spore Germination.Trends Microbiol. 2020 Sep;28(9):744-752. doi: 10.1016/j.tim.2020.03.004. Epub 2020 Apr 23. Trends Microbiol. 2020. PMID: 32781028 Review.
-
Clostridioides difficile Spore Formation and Germination: New Insights and Opportunities for Intervention.Annu Rev Microbiol. 2020 Sep 8;74:545-566. doi: 10.1146/annurev-micro-011320-011321. Annu Rev Microbiol. 2020. PMID: 32905755 Review.
Cited by
-
Observations on research with spores of Bacillales and Clostridiales species.J Appl Microbiol. 2019 Feb;126(2):348-358. doi: 10.1111/jam.14067. Epub 2018 Sep 27. J Appl Microbiol. 2019. PMID: 30106202 Free PMC article. Review.
-
The requirement for co-germinants during Clostridium difficile spore germination is influenced by mutations in yabG and cspA.PLoS Pathog. 2019 Apr 3;15(4):e1007681. doi: 10.1371/journal.ppat.1007681. eCollection 2019 Apr. PLoS Pathog. 2019. PMID: 30943268 Free PMC article.
-
Unique growth and morphology properties of Clade 5 Clostridioides difficile strains revealed by single-cell time-lapse microscopy.PLoS Pathog. 2025 May 21;21(5):e1013155. doi: 10.1371/journal.ppat.1013155. eCollection 2025 May. PLoS Pathog. 2025. PMID: 40397889 Free PMC article.
-
A sporulation signature protease is required for assembly of the spore surface layers, germination and host colonization in Clostridioides difficile.PLoS Pathog. 2023 Nov 13;19(11):e1011741. doi: 10.1371/journal.ppat.1011741. eCollection 2023 Nov. PLoS Pathog. 2023. PMID: 37956166 Free PMC article.
-
Functional redundancy between penicillin-binding proteins during asymmetric cell division in Clostridioides difficile.bioRxiv [Preprint]. 2025 Mar 7:2024.09.26.615255. doi: 10.1101/2024.09.26.615255. bioRxiv. 2025. PMID: 39386573 Free PMC article. Preprint.
References
-
- Warny M, Pepin J, Fang A, Killgore G, Thompson A, et al. (2005) Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366: 1079–1084. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

