Biochemical analysis of the multifunctional vaccinia mRNA capping enzyme encoded by a temperature sensitive virus mutant
- PMID: 26496697
- PMCID: PMC4679637
- DOI: 10.1016/j.virol.2015.10.011
Biochemical analysis of the multifunctional vaccinia mRNA capping enzyme encoded by a temperature sensitive virus mutant
Abstract
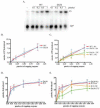
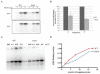
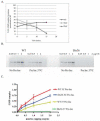
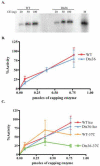
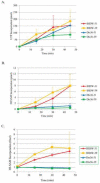
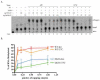
Prior biochemical analysis of the heterodimeric vaccinia virus mRNA capping enzyme suggests roles not only in mRNA capping but also in early viral gene transcription termination and intermediate viral gene transcription initiation. Prior phenotypic characterization of Dts36, a temperature sensitive virus mutant affecting the large subunit of the capping enzyme was consistent with the multifunctional roles of the capping enzyme in vivo. We report a biochemical analysis of the capping enzyme encoded by Dts36. Of the three enzymatic activities required for mRNA capping, the guanylyltransferase and methyltransferase activities are compromised while the triphosphatase activity and the D12 subunit interaction are unaffected. The mutant enzyme is also defective in stimulating early gene transcription termination and intermediate gene transcription initiation in vitro. These results confirm that the vaccinia virus mRNA capping enzyme functions not only in mRNA capping but also early gene transcription termination and intermediate gene transcription initiation in vivo.
Keywords: Capping enzyme; Transcription initiation; Transcription termination; Vaccinia virus; mRNA capping.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures




Similar articles
-
Analysis of a temperature-sensitive vaccinia virus mutant in the viral mRNA capping enzyme isolated by clustered charge-to-alanine mutagenesis and transient dominant selection.Virology. 1997 Nov 24;238(2):391-409. doi: 10.1006/viro.1997.8820. Virology. 1997. PMID: 9400612
-
A yeast-based genetic system for functional analysis of viral mRNA capping enzymes.J Virol. 2000 Jun;74(12):5486-94. doi: 10.1128/jvi.74.12.5486-5494.2000. J Virol. 2000. PMID: 10823853 Free PMC article.
-
Structure-function analysis of the triphosphatase component of vaccinia virus mRNA capping enzyme.J Virol. 1997 Dec;71(12):9837-43. doi: 10.1128/JVI.71.12.9837-9843.1997. J Virol. 1997. PMID: 9371657 Free PMC article.
-
Studying vaccinia virus RNA processing in vitro.Methods Mol Biol. 2004;269:151-68. doi: 10.1385/1-59259-789-0:151. Methods Mol Biol. 2004. PMID: 15114015 Review.
-
Regulation of viral transcription elongation and termination during vaccinia virus infection.Biochim Biophys Acta. 2002 Sep 13;1577(2):325-36. doi: 10.1016/s0167-4781(02)00461-x. Biochim Biophys Acta. 2002. PMID: 12213661 Review.
References
-
- Broyles SS. Vaccinia virus transcription. J.Gen.Virol. 2003;84:2293–2303. - PubMed
-
- Christen LM, Sanders M, Wiler C, Niles EG. Vaccinia virus nucleoside triphosphate phosphohydrolase I is an essential viral early gene transcription termination factor. Virology. 1998;245:360–371. - PubMed
-
- Condit RC, Lewis JI, Quinn M, Christen LM, Niles EG. Use of lysolecithin-permeabilized infected-cell extracts to investigate the in vitro biochemical phenotypes of poxvirus ts mutations altered in viral transcription activity. Virology. 1996;218:169–180. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
