CstF-64 and 3'-UTR cis-element determine Star-PAP specificity for target mRNA selection by excluding PAPα
- PMID: 26496945
- PMCID: PMC4737136
- DOI: 10.1093/nar/gkv1074
CstF-64 and 3'-UTR cis-element determine Star-PAP specificity for target mRNA selection by excluding PAPα
Erratum in
-
CstF-64 and 3'-UTR cis-element determine Star-PAP specificity for target mRNA selection by excluding PAPα.Nucleic Acids Res. 2019 Feb 20;47(3):1598. doi: 10.1093/nar/gky1248. Nucleic Acids Res. 2019. PMID: 30566625 Free PMC article. No abstract available.
Abstract
Almost all eukaryotic mRNAs have a poly (A) tail at the 3'-end. Canonical PAPs (PAPα/γ) polyadenylate nuclear pre-mRNAs. The recent identification of the non-canonical Star-PAP revealed specificity of nuclear PAPs for pre-mRNAs, yet the mechanism how Star-PAP selects mRNA targets is still elusive. Moreover, how Star-PAP target mRNAs having canonical AAUAAA signal are not regulated by PAPα is unclear. We investigate specificity mechanisms of Star-PAP that selects pre-mRNA targets for polyadenylation. Star-PAP assembles distinct 3'-end processing complex and controls pre-mRNAs independent of PAPα. We identified a Star-PAP recognition nucleotide motif and showed that suboptimal DSE on Star-PAP target pre-mRNA 3'-UTRs inhibit CstF-64 binding, thus preventing PAPα recruitment onto it. Altering 3'-UTR cis-elements on a Star-PAP target pre-mRNA can switch the regulatory PAP from Star-PAP to PAPα. Our results suggest a mechanism of poly (A) site selection that has potential implication on the regulation of alternative polyadenylation.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
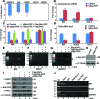

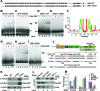
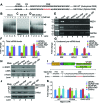
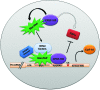
Similar articles
-
Poly(A) polymerase (PAP) diversity in gene expression--star-PAP vs canonical PAP.FEBS Lett. 2014 Jun 27;588(14):2185-97. doi: 10.1016/j.febslet.2014.05.029. Epub 2014 May 27. FEBS Lett. 2014. PMID: 24873880 Free PMC article. Review.
-
The poly A polymerase Star-PAP controls 3'-end cleavage by promoting CPSF interaction and specificity toward the pre-mRNA.EMBO J. 2010 Dec 15;29(24):4132-45. doi: 10.1038/emboj.2010.287. Epub 2010 Nov 19. EMBO J. 2010. PMID: 21102410 Free PMC article.
-
Star-PAP controlled alternative polyadenylation coupled poly(A) tail length regulates protein expression in hypertrophic heart.Nucleic Acids Res. 2019 Nov 18;47(20):10771-10787. doi: 10.1093/nar/gkz875. Nucleic Acids Res. 2019. PMID: 31598705 Free PMC article.
-
Distinct regulation of alternative polyadenylation and gene expression by nuclear poly(A) polymerases.Nucleic Acids Res. 2017 Sep 6;45(15):8930-8942. doi: 10.1093/nar/gkx560. Nucleic Acids Res. 2017. PMID: 28911096 Free PMC article.
-
The novel poly(A) polymerase Star-PAP is a signal-regulated switch at the 3'-end of mRNAs.Adv Biol Regul. 2013 Jan;53(1):64-76. doi: 10.1016/j.jbior.2012.10.004. Epub 2012 Oct 13. Adv Biol Regul. 2013. PMID: 23306079 Free PMC article. Review.
Cited by
-
CSTF2-impeded innate αβ T cell infiltration and activation exacerbate immune evasion of pancreatic cancer.Cell Death Differ. 2025 May;32(5):973-988. doi: 10.1038/s41418-025-01464-0. Epub 2025 Feb 19. Cell Death Differ. 2025. PMID: 39972059
-
CPSF6 links alternative polyadenylation to metabolism adaption in hepatocellular carcinoma progression.J Exp Clin Cancer Res. 2021 Mar 1;40(1):85. doi: 10.1186/s13046-021-01884-z. J Exp Clin Cancer Res. 2021. PMID: 33648552 Free PMC article.
-
Alternative cleavage and polyadenylation: key regulatory mechanisms in health and disease.RNA Biol. 2025 Dec;22(1):1-33. doi: 10.1080/15476286.2025.2529033. Epub 2025 Jul 9. RNA Biol. 2025. PMID: 40613493 Free PMC article. Review.
-
Nuclear Phosphatidylinositol-Phosphate Type I Kinase α-Coupled Star-PAP Polyadenylation Regulates Cell Invasion.Mol Cell Biol. 2018 Feb 12;38(5):e00457-17. doi: 10.1128/MCB.00457-17. Print 2018 Mar 1. Mol Cell Biol. 2018. PMID: 29203642 Free PMC article.
-
Terminal nucleotidyl transferases (TENTs) in mammalian RNA metabolism.Philos Trans R Soc Lond B Biol Sci. 2018 Nov 5;373(1762):20180162. doi: 10.1098/rstb.2018.0162. Philos Trans R Soc Lond B Biol Sci. 2018. PMID: 30397099 Free PMC article. Review.
References
-
- Colgan D.F., Manley J.L. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. - PubMed
-
- Proudfoot N. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr. Opin. Cell Biol. 2004;16:272–278. - PubMed
-
- Edmonds M. A history of poly A sequences: from formation to factors to function. Prog. Nucleic Acid Res. Mol. Biol. 2002;71:285–389. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

