Transient overexpression of DNA adenine methylase enables efficient and mobile genome engineering with reduced off-target effects
- PMID: 26496947
- PMCID: PMC4770203
- DOI: 10.1093/nar/gkv1090
Transient overexpression of DNA adenine methylase enables efficient and mobile genome engineering with reduced off-target effects
Abstract
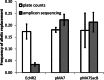
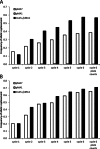
Homologous recombination of single-stranded oligonucleotides is a highly efficient process for introducing precise mutations into the genome of E. coli and other organisms when mismatch repair (MMR) is disabled. This can result in the rapid accumulation of off-target mutations that can mask desired phenotypes, especially when selections need to be employed following the generation of combinatorial libraries. While the use of inducible mutator phenotypes or other MMR evasion tactics have proven useful, reported methods either require non-mobile genetic modifications or costly oligonucleotides that also result in reduced efficiencies of replacement. Therefore a new system was developed, Transient Mutator Multiplex Automated Genome Engineering (TM-MAGE), that solves problems encountered in other methods for oligonucleotide-mediated recombination. TM-MAGE enables nearly equivalent efficiencies of allelic replacement to the use of strains with fully disabled MMR and with an approximately 12- to 33-fold lower off-target mutation rate. Furthermore, growth temperatures are not restricted and a version of the plasmid can be readily removed by sucrose counterselection. TM-MAGE was used to combinatorially reconstruct mutations found in evolved salt-tolerant strains, enabling the identification of causative mutations and isolation of strains with up to 75% increases in growth rate and greatly reduced lag times in 0.6 M NaCl.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





Similar articles
-
Merlin: Computer-Aided Oligonucleotide Design for Large Scale Genome Engineering with MAGE.ACS Synth Biol. 2016 Jun 17;5(6):452-8. doi: 10.1021/acssynbio.5b00219. Epub 2016 May 11. ACS Synth Biol. 2016. PMID: 27054880
-
Highly efficient deletion method for the engineering of plasmid DNA with single-stranded oligonucleotides.Biotechniques. 2008 Feb;44(2):217-20, 222, 224. doi: 10.2144/000112684. Biotechniques. 2008. PMID: 18330349
-
Conditional DNA repair mutants enable highly precise genome engineering.Nucleic Acids Res. 2014 Apr;42(8):e62. doi: 10.1093/nar/gku105. Epub 2014 Feb 5. Nucleic Acids Res. 2014. PMID: 24500200 Free PMC article.
-
Genome-scale genetic engineering in Escherichia coli.Biotechnol Adv. 2013 Nov;31(6):804-10. doi: 10.1016/j.biotechadv.2013.04.003. Epub 2013 Apr 24. Biotechnol Adv. 2013. PMID: 23624241 Review.
-
[Recombineering and its application].Yi Chuan Xue Bao. 2003 Oct;30(10):983-8. Yi Chuan Xue Bao. 2003. PMID: 14669518 Review. Chinese.
Cited by
-
Unpredictability of the Fitness Effects of Antimicrobial Resistance Mutations Across Environments in Escherichia coli.Mol Biol Evol. 2024 May 3;41(5):msae086. doi: 10.1093/molbev/msae086. Mol Biol Evol. 2024. PMID: 38709811 Free PMC article.
-
Dominant resistance and negative epistasis can limit the co-selection of de novo resistance mutations and antibiotic resistance genes.Nat Commun. 2020 Mar 5;11(1):1199. doi: 10.1038/s41467-020-15080-8. Nat Commun. 2020. PMID: 32139686 Free PMC article.
-
Automated growth rate determination in high-throughput microbioreactor systems.BMC Res Notes. 2017 Nov 25;10(1):617. doi: 10.1186/s13104-017-2945-6. BMC Res Notes. 2017. PMID: 29178966 Free PMC article.
-
CRISPR-TSKO: A Technique for Efficient Mutagenesis in Specific Cell Types, Tissues, or Organs in Arabidopsis.Plant Cell. 2019 Dec;31(12):2868-2887. doi: 10.1105/tpc.19.00454. Epub 2019 Sep 27. Plant Cell. 2019. PMID: 31562216 Free PMC article.
-
An sRNA Screen for Reversal of Quinolone Resistance in Escherichia coli.G3 (Bethesda). 2020 Jan 7;10(1):79-88. doi: 10.1534/g3.119.400199. G3 (Bethesda). 2020. PMID: 31744901 Free PMC article.
References
-
- Thomason L., Court D.L., Bubunenko M., Constantino N., Wilson H., Datta S., Oppenheim A. Recombineering: genetic engineering in bacteria using homologous recombination. 2007 Unit1.16. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

