The CreC Regulator of Escherichia coli, a New Target for Metabolic Manipulations
- PMID: 26497466
- PMCID: PMC4702642
- DOI: 10.1128/AEM.02984-15
The CreC Regulator of Escherichia coli, a New Target for Metabolic Manipulations
Abstract
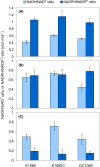
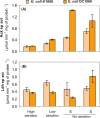
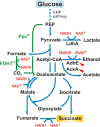
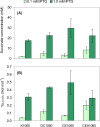
The CreBC (carbon source-responsive) two-component regulation system of Escherichia coli affects a number of functions, including intermediary carbon catabolism. The impacts of different creC mutations (a ΔcreC mutant and a mutant carrying the constitutive creC510 allele) on bacterial physiology were analyzed in glucose cultures under three oxygen availability conditions. Differences in the amounts of extracellular metabolites produced were observed in the null mutant compared to the wild-type strain and the mutant carrying creC510 and shown to be affected by oxygen availability. The ΔcreC strain secreted more formate, succinate, and acetate but less lactate under low aeration. These metabolic changes were associated with differences in AckA and LdhA activities, both of which were affected by CreC. Measurement of the NAD(P)H/NAD(P)(+) ratios showed that the creC510 strain had a more reduced intracellular redox state, while the opposite was observed for the ΔcreC mutant, particularly under intermediate oxygen availability conditions, indicating that CreC affects redox balance. The null mutant formed more succinate than the wild-type strain under both low aeration and no aeration. Overexpression of the genes encoding phosphoenolpyruvate carboxylase from E. coli and a NADH-forming formate dehydrogenase from Candida boidinii in the ΔcreC mutant further increased the yield of succinate on glucose. Interestingly, the elimination of ackA and adhE did not significantly improve the production of succinate. The diverse metabolic effects of this regulator on the central biochemical network of E. coli make it a good candidate for metabolic-engineering manipulations to enhance the formation of bioproducts, such as succinate.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
Similar articles
-
Microbial Cell Factories à la Carte: Elimination of Global Regulators Cra and ArcA Generates Metabolic Backgrounds Suitable for the Synthesis of Bioproducts in Escherichia coli.Appl Environ Microbiol. 2018 Sep 17;84(19):e01337-18. doi: 10.1128/AEM.01337-18. Print 2018 Oct 1. Appl Environ Microbiol. 2018. PMID: 30030227 Free PMC article.
-
High-yield anaerobic succinate production by strategically regulating multiple metabolic pathways based on stoichiometric maximum in Escherichia coli.Microb Cell Fact. 2016 Aug 12;15(1):141. doi: 10.1186/s12934-016-0536-1. Microb Cell Fact. 2016. PMID: 27520031 Free PMC article.
-
Pyruvate dehydrogenase complex regulator (PdhR) gene deletion boosts glucose metabolism in Escherichia coli under oxygen-limited culture conditions.J Biosci Bioeng. 2017 Apr;123(4):437-443. doi: 10.1016/j.jbiosc.2016.11.004. Epub 2016 Dec 20. J Biosci Bioeng. 2017. PMID: 28007420
-
Current advances of succinate biosynthesis in metabolically engineered Escherichia coli.Biotechnol Adv. 2017 Dec;35(8):1040-1048. doi: 10.1016/j.biotechadv.2017.09.007. Epub 2017 Sep 20. Biotechnol Adv. 2017. PMID: 28939498 Review.
-
Succinate production in Escherichia coli.Biotechnol J. 2012 Feb;7(2):213-24. doi: 10.1002/biot.201100061. Epub 2011 Sep 20. Biotechnol J. 2012. PMID: 21932253 Free PMC article. Review.
Cited by
-
Microbial Cell Factories à la Carte: Elimination of Global Regulators Cra and ArcA Generates Metabolic Backgrounds Suitable for the Synthesis of Bioproducts in Escherichia coli.Appl Environ Microbiol. 2018 Sep 17;84(19):e01337-18. doi: 10.1128/AEM.01337-18. Print 2018 Oct 1. Appl Environ Microbiol. 2018. PMID: 30030227 Free PMC article.
-
Genome Insights of the Plant-Growth Promoting Bacterium Cronobacter muytjensii JZ38 With Volatile-Mediated Antagonistic Activity Against Phytophthora infestans.Front Microbiol. 2020 Mar 11;11:369. doi: 10.3389/fmicb.2020.00369. eCollection 2020. Front Microbiol. 2020. PMID: 32218777 Free PMC article.
-
Comparative genomic analysis of the Hafnia genus reveals an explicit evolutionary relationship between the species alvei and paralvei and provides insights into pathogenicity.BMC Genomics. 2019 Oct 23;20(1):768. doi: 10.1186/s12864-019-6123-1. BMC Genomics. 2019. PMID: 31646960 Free PMC article.
-
Refactoring the Embden-Meyerhof-Parnas Pathway as a Whole of Portable GlucoBricks for Implantation of Glycolytic Modules in Gram-Negative Bacteria.ACS Synth Biol. 2017 May 19;6(5):793-805. doi: 10.1021/acssynbio.6b00230. Epub 2017 Feb 9. ACS Synth Biol. 2017. PMID: 28121421 Free PMC article.
-
A High-Throughput Method for Identifying Novel Genes That Influence Metabolic Pathways Reveals New Iron and Heme Regulation in Pseudomonas aeruginosa.mSystems. 2021 Feb 2;6(1):e00933-20. doi: 10.1128/mSystems.00933-20. mSystems. 2021. PMID: 33531406 Free PMC article.
References
-
- Bettenbrock K, Bai H, Ederer M, Green J, Hellingwerf KJ, Holcombe M, Kunz S, Rolfe MD, Sanguinetti G, Sawodny O, Sharma P, Steinsiek S, Poole RK. 2014. Towards a systems level understanding of the oxygen response of Escherichia coli.. Adv Microb Physiol 64:65–114. doi:10.1016/B978-0-12-800143-1.00002-6. - DOI - PubMed
-
- Rolfe MD, Ter Beek A, Graham AI, Trotter EW, Asif HMS, Sanguinetti G, Teixeira de Mattos MJ, Poole RK, Green J. 2011. Transcript profiling and inference of Escherichia coli K-12 ArcA activity across the range of physiologically relevant oxygen concentrations. J Biol Chem 286:10147–10154. doi:10.1074/jbc.M110.211144. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

