Impact of efflux in the development of multidrug resistance phenotypes in Staphylococcus aureus
- PMID: 26498754
- PMCID: PMC4619429
- DOI: 10.1186/s12866-015-0572-8
Impact of efflux in the development of multidrug resistance phenotypes in Staphylococcus aureus
Abstract
Background: Efflux has been recognized as a resistance mechanism to antimicrobials in Staphylococcus aureus; however its role on the development of clinically relevant resistance is still poorly characterized. This study aimed to examine the impact of efflux on development of resistance to fluoroquinolones and other antimicrobials in S. aureus strains representing relevant phenotypes in terms of antibiotic susceptibility and efflux activity.
Methods: Two closely related methicillin- and ciprofloxacin-resistant Staphylococcus aureus clinical strains, with different efflux capacity and the pan-susceptible strain ATCC25923 were exposed to constant concentrations of the efflux pump (EP) substrates ciprofloxacin, ethidium bromide and cetrimide. Parental and exposed strains were tested regarding their susceptibility towards antibiotics, biocides and ethidium bromide, efflux capacity and levels of EP gene expression. Occurrence of resistance-associated mutations was screened by sequencing.
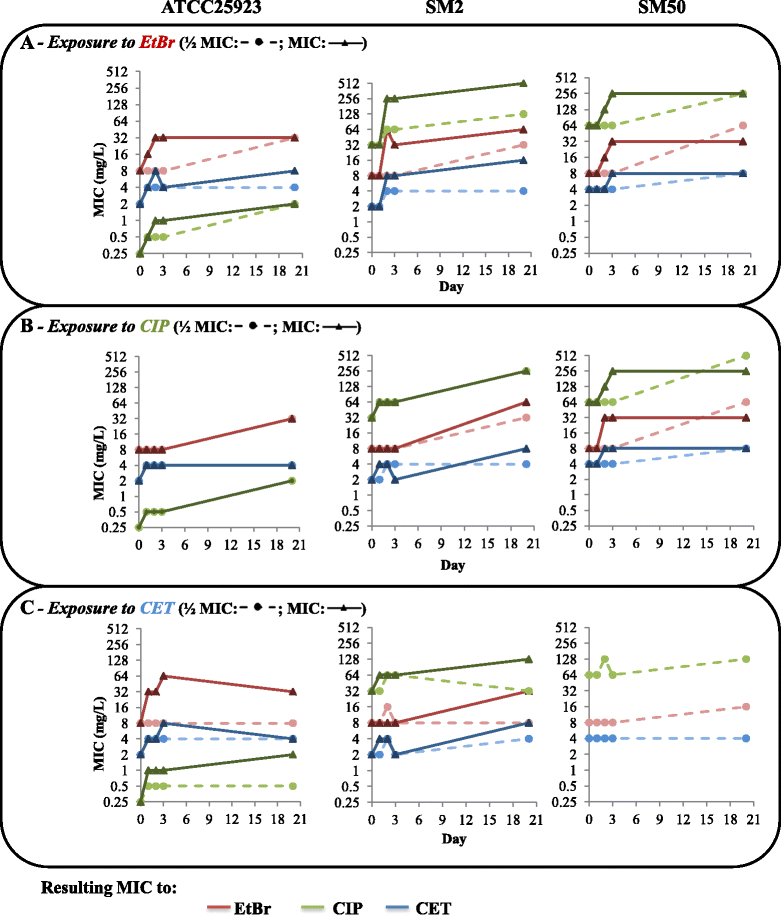
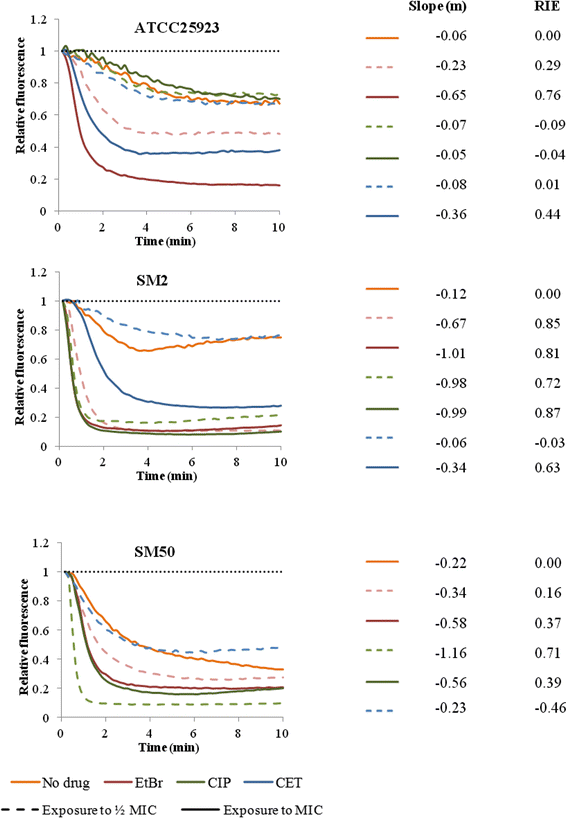
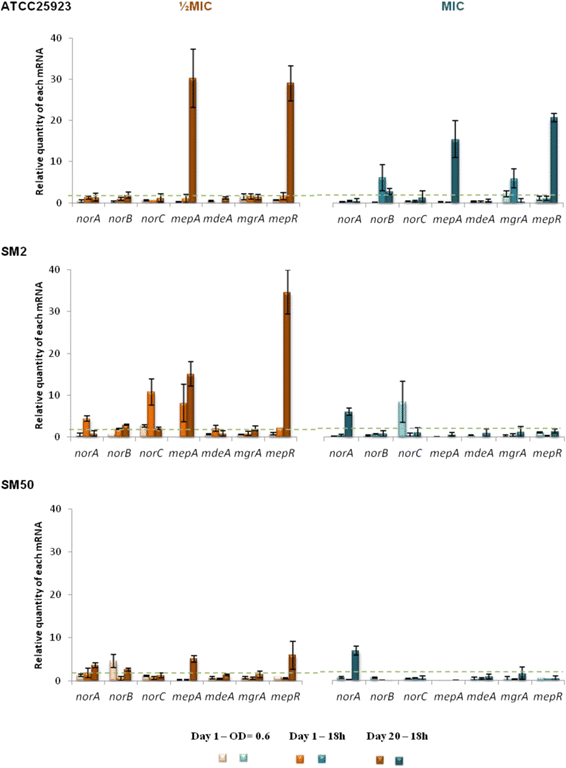
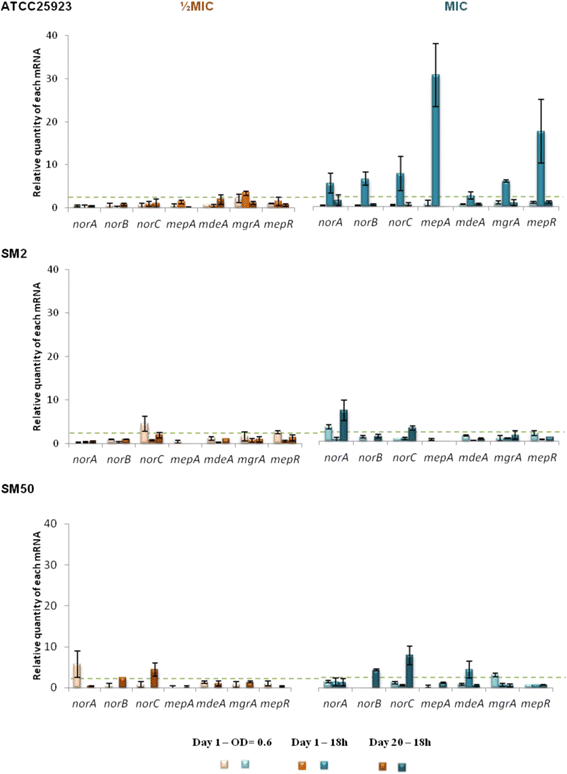
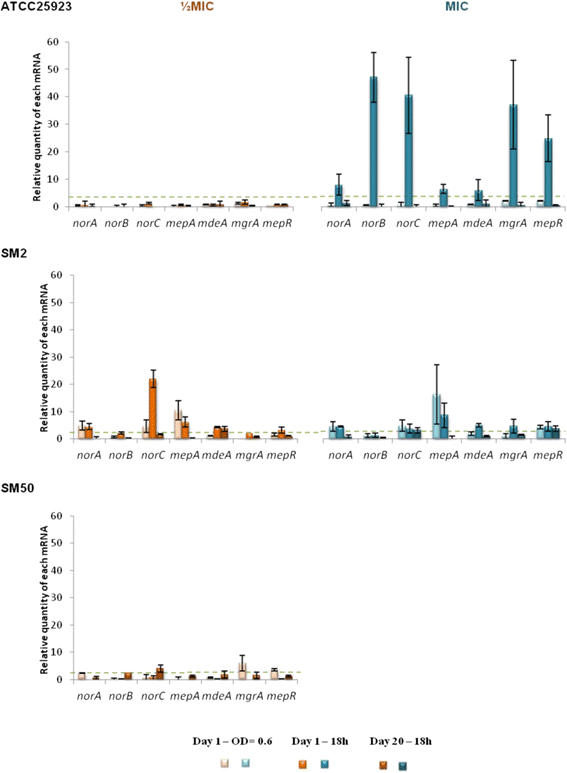
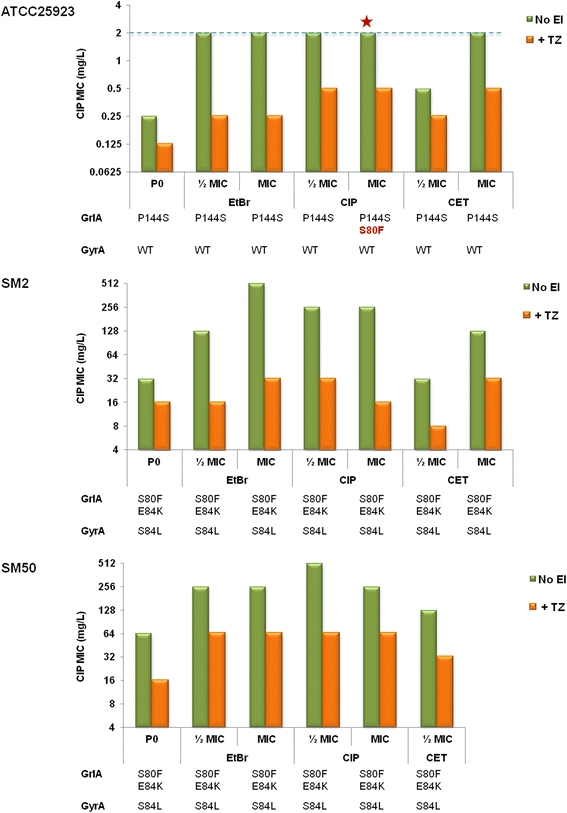
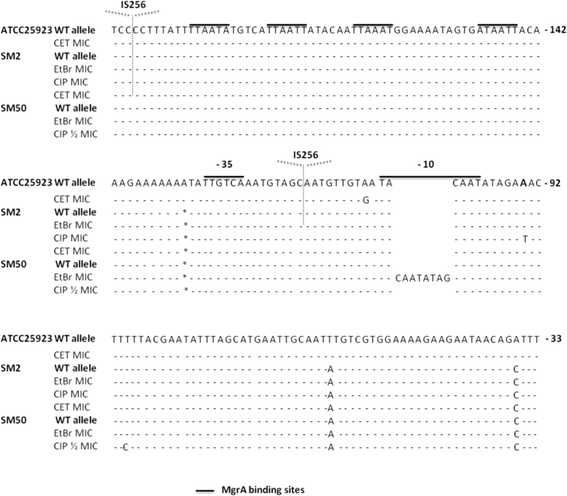
Results: Multidrug resistance phenotypes emerged upon exposure, independently of the substrate or its concentration, which were correlated with increased efflux capacity of the exposed strains. The temporal pattern of EP gene expression disclosed an early-response with high expression of several genes, followed by a late-response, characterized by overexpression of specific genes. The overall cell response was more pronounced for strains with an initial basal efflux activity. Remarkably, detection of the IS256 element in the promoter regions of mgrA and norA, in some cases associated with increased gene expression, suggests that these genes may be hot spots for IS256 insertion events. The results obtained with exposure of ATCC25923 to ciprofloxacin were particularly striking, revealing a step-wise development of fluoroquinolone resistance, with a first efflux-mediated response, followed by the occurrence of a mutation in grlA that resulted in phenotypic resistance. Additionally, challenge by non-fluoroquinolone agents, particularly cetrimide, promoted cross resistance to fluoroquinolones, revealing the potential role of biocides as selective pressure for the emergence of resistance to these antibiotics.
Conclusions: This study reveals efflux as a significant component of S. aureus resistance to fluoroquinolones and biocides and as a primary mechanism to withstand stress imposed by antimicrobials. This efflux-mediated response can result in the emergence of multidrug resistance in healthcare environments and should be taken into account in the management of this major pathogen.
Figures
References
-
- European Centre for Disease Prevention and Control (ECDC). Antimicrobial Resistance Surveillance in Europe 2013. Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net), ECDC: Stockholm, Sweden, 2014. Available: http://www.ecdc.europa.eu/en/publications/Publications/antimicrobial-res....
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

