Leveraging local ancestry to detect gene-gene interactions in genome-wide data
- PMID: 26498930
- PMCID: PMC4619349
- DOI: 10.1186/s12863-015-0283-z
Leveraging local ancestry to detect gene-gene interactions in genome-wide data
Abstract
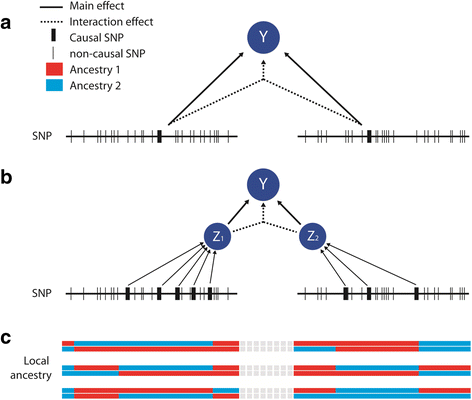
Background: Although genome-wide association studies have successfully identified thousands of variants associated to complex traits, these variants only explain a small amount of the entire heritability of the trait. Gene-gene interactions have been proposed as a source to explain a significant percentage of the missing heritability. However, detecting gene-gene interactions has proven to be very difficult due to computational and statistical challenges. The vast number of possible interactions that can be tested induces very stringent multiple hypotheses corrections that limit the power of detection. These issues have been mostly highlighted for the identification of pairwise effects and are even more challenging when addressing higher order interaction effects. In this work we explore the use of local ancestry in recently admixed individuals to find signals of gene-gene interaction on human traits and diseases.
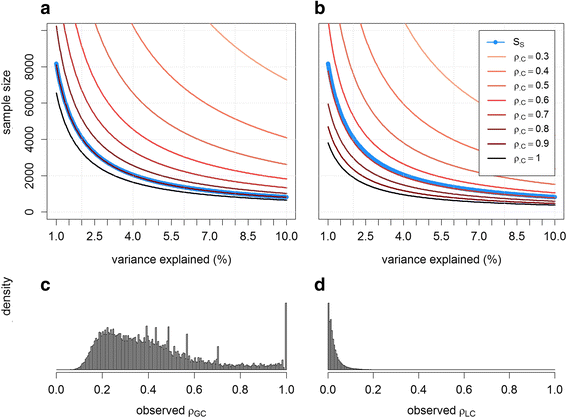
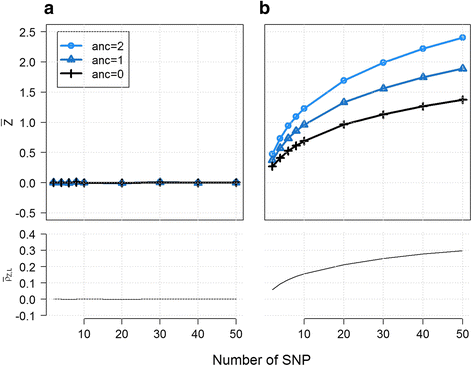
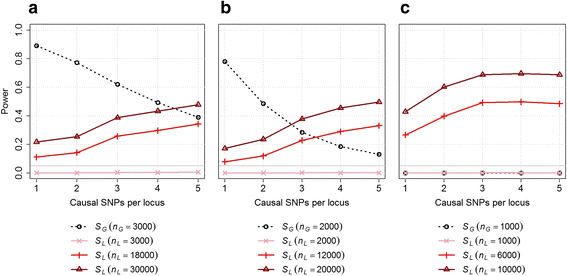
Results: We introduce statistical methods that leverage the correlation between local ancestry and the hidden unknown causal variants to find distant gene-gene interactions. We show that the power of this test increases with the number of causal variants per locus and the degree of differentiation of these variants between the ancestral populations. Overall, our simulations confirm that local ancestry can be used to detect gene-gene interactions, solving the computational bottleneck. When compared to a single nucleotide polymorphism (SNP)-based interaction screening of the same sample size, the power of our test was lower on all settings we considered. However, accounting for the dramatic increase in sample size that can be achieve when genotyping only a set of ancestry informative markers instead of the whole genome, we observe substantial gain in power in several scenarios.
Conclusion: Local ancestry-based interaction tests offer a new path to the detection of gene-gene interaction effects. It would be particularly useful in scenarios where multiple differentiated variants at the interacting loci act in a synergistic manner.
Figures
References
-
- Powell R, Davidson D, Divers J, Manichaikul A, Carr JJ, Detrano R, Hoffman EA, Jiang R, Kronmal RA, Liu K, et al. Genetic ancestry and the relationship of cigarette smoking to lung function and per cent emphysema in four race/ethnic groups: a cross-sectional study. Thorax. 2013;68(7):634–642. doi: 10.1136/thoraxjnl-2012-202116. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

