Extracting Accurate Precursor Information for Tandem Mass Spectra by RawConverter
- PMID: 26499134
- PMCID: PMC4777630
- DOI: 10.1021/acs.analchem.5b02721
Extracting Accurate Precursor Information for Tandem Mass Spectra by RawConverter
Abstract
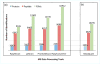
Extraction of data from the proprietary RAW files generated by Thermo Fisher mass spectrometers is the primary step for subsequent data analysis. High resolution and high mass accuracy data obtained by state-of-the-art mass spectrometers (e.g., Orbitraps) can significantly improve both peptide/protein identification and quantification. We developed RawConverter, a stand-alone software tool, to improve data extraction on RAW files from high-resolution Thermo Fisher mass spectrometers. RawConverter extracts full scan and MS(n) data from RAW files like its predecessor RawXtract; most importantly, it associates the accurate precursor mass-to-charge (m/z) value with the tandem mass spectrum. RawConverter accepts RAW data generated by either data-dependent acquisition (DDA) or data-independent acquisition (DIA). It generates output into MS1/MS2/MS3, MGF, or mzXML file formats, which fulfills the format requirements for most data identification and quantification tools. Using the tandem mass spectra extracted by RawConverter with corrected m/z values, 32.8%, 27.1%, and 84.1%, peptide spectra matches (PSMs) produce 17.4% (13.0%), 14.4% (11.5%), and 45.7% (36.2%) more peptide (protein) identifications than ProteoWizard, pXtract, and RawXtract, respectively. RawConverter is implemented in C# and is freely accessible at http://fields.scripps.edu/rawconv.
Figures

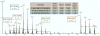

References
-
- McDonald WH, Tabb DL, Sadygov RG, MacCoss MJ, Venable J, Graumann J, Johnson JR, Cociorva D, Yates JR., III Rapid Commun Mass Spectrom. 2004;18:2162–2168. - PubMed
-
- Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, Hoff K, Kessner D, Tasman N, Shulman N, Frewen B, Baker TA, Brusniak M, Paulse C, Creasy D, Flashner L, Kani K, Moulding C, Seymour SL, Nuwaysir LM, Lefebvre B, Kuhlmann F, Roark J, Rainer P, Detlev S, Hemenway T, Huhmer A, Langridge J, Connolly B, Chadick T, Holly K, Eckels J, Deutsch EW, Moritz RL, Katz JE, Agus DB, MacCoss MJ, Tabb DL, Mallick P. Nat Biotechnol. 2012;30:918–920. - PMC - PubMed
-
- Yuan ZF, Liu C, Wang HP, Sun RX, Fu Y, Zhang JF, Wang LH, Chi H, Li Y, Xiu LY, Wang WP, He SM. Proteomics. 2012;12(2):226–235. - PubMed
-
- Hao P, Ren Y, Tam JP, Sze SK. J Proteome Res. 2013;12(12):5548–5557. - PubMed
-
- Nassar AF, Hollenberg P, Scatina J. Drug metabolism handbook: concepts and applications. John Wiley and Sons; Hoboken, New Jersey: 2009. p. 216.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

