Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes
- PMID: 26499245
- PMCID: PMC4664323
- DOI: 10.1073/pnas.1518552112
Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes
Abstract
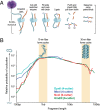
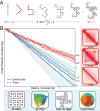
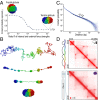
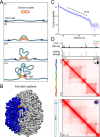
We recently used in situ Hi-C to create kilobase-resolution 3D maps of mammalian genomes. Here, we combine these maps with new Hi-C, microscopy, and genome-editing experiments to study the physical structure of chromatin fibers, domains, and loops. We find that the observed contact domains are inconsistent with the equilibrium state for an ordinary condensed polymer. Combining Hi-C data and novel mathematical theorems, we show that contact domains are also not consistent with a fractal globule. Instead, we use physical simulations to study two models of genome folding. In one, intermonomer attraction during polymer condensation leads to formation of an anisotropic "tension globule." In the other, CCCTC-binding factor (CTCF) and cohesin act together to extrude unknotted loops during interphase. Both models are consistent with the observed contact domains and with the observation that contact domains tend to form inside loops. However, the extrusion model explains a far wider array of observations, such as why loops tend not to overlap and why the CTCF-binding motifs at pairs of loop anchors lie in the convergent orientation. Finally, we perform 13 genome-editing experiments examining the effect of altering CTCF-binding sites on chromatin folding. The convergent rule correctly predicts the affected loops in every case. Moreover, the extrusion model accurately predicts in silico the 3D maps resulting from each experiment using only the location of CTCF-binding sites in the WT. Thus, we show that it is possible to disrupt, restore, and move loops and domains using targeted mutations as small as a single base pair.
Keywords: CRISPR; CTCF; chromatin loops; genome architecture; molecular dynamics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

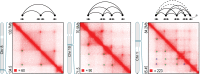

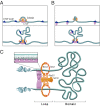
Comment in
-
Predictive model of 3D domain formation via CTCF-mediated extrusion.Proc Natl Acad Sci U S A. 2015 Nov 24;112(47):14404-5. doi: 10.1073/pnas.1519849112. Epub 2015 Nov 13. Proc Natl Acad Sci U S A. 2015. PMID: 26567152 Free PMC article. No abstract available.
References
-
- Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98(3):285–294. - PubMed
-
- Hansen JC. Conformational dynamics of the chromatin fiber in solution: Determinants, mechanisms, and functions. Annu Rev Biophys Biomol Struct. 2002;31(1):361–392. - PubMed
-
- Hartwig M. The size of independently supercoiled domains in nuclear DNA from normal human lymphocytes and leukemic lymphoblasts. Biochim Biophys Acta. 1982;698(2):214–217. - PubMed
-
- Zehnbauer BA, Vogelstein B. Supercoiled loops and the organization of replication and transcription in eukaryotes. BioEssays. 1985;2(2):52–54.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

