Pleiotropic Functions of Tumor Suppressor WWOX in Normal and Cancer Cells
- PMID: 26499798
- PMCID: PMC4692203
- DOI: 10.1074/jbc.R115.676346
Pleiotropic Functions of Tumor Suppressor WWOX in Normal and Cancer Cells
Abstract
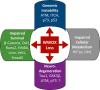
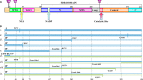
WW domain-containing oxidoreductase (WWOX), originally marked as a likely tumor suppressor gene, has over the years become recognized for its role in a much wider range of cellular activities. Phenotypic effects displayed in animal studies, along with resolution of WWOX's architecture, fold, and binding partners, point to the protein's multifaceted biological functions. Results from a series of complementary experiments seem to indicate WWOX's involvement in metabolic regulation. More recently, clinical studies involving cases of severe encephalopathy suggest that WWOX also plays a part in controlling CNS development, further expanding our understanding of the breadth and complexity of WWOX behavior. Here we present a short overview of the various approaches taken to study this dynamic gene, emphasizing the most recent findings regarding WWOX's metabolic- and CNS-associated functions and their underlying molecular basis.
Keywords: SDR domain; WW domain; WWOX; animal model; animal models; cell metabolism; central nervous system (CNS); common fragile site; epilepsy; protein-protein interaction; tumor suppressor; tumor suppressor gene.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

References
-
- Bednarek A. K., Laflin K. J., Daniel R. L., Liao Q., Hawkins K. A., and Aldaz C. M. (2000) WWOX, a novel WW domain-containing protein mapping to human chromosome 16q23.3-24.1, a region frequently affected in breast cancer. Cancer Res. 60, 2140–2145 - PubMed
-
- Ried K., Finnis M., Hobson L., Mangelsdorf M., Dayan S., Nancarrow J. K., Woollatt E., Kremmidiotis G., Gardner A., Venter D., Baker E., and Richards R. I. (2000) Common chromosomal fragile site FRA16D sequence: identification of the FOR gene spanning FRA16D and homozygous deletions and translocation breakpoints in cancer cells. Hum. Mol. Genet. 9, 1651–1663 - PubMed
-
- Durkin S. G., and Glover T. W. (2007) Chromosome fragile sites. Annu. Rev. Genet. 41, 169–192 - PubMed
-
- Beroukhim R., Mermel C. H., Porter D., Wei G., Raychaudhuri S., Donovan J., Barretina J., Boehm J. S., Dobson J., Urashima M., Mc Henry K. T., Pinchback R. M., Ligon A. H., Cho Y. J., Haery L., Greulich H., Reich M., Winckler W., Lawrence M. S., Weir B. A., Tanaka K. E., Chiang D. Y., Bass A. J., Loo A., Hoffman C., Prensner J., Liefeld T., Gao Q., Yecies D., Signoretti S., Maher E., Kaye F. J., Sasaki H., Tepper J. E., Fletcher J. A., Tabernero J., Baselga J., Tsao M. S., Demichelis F., Rubin M. A., Janne P. A., Daly M. J., Nucera C., Levine R. L., Ebert B. L., Gabriel S., Rustgi A. K., Antonescu C. R., Ladanyi M., Letai A., Garraway L. A., Loda M., Beer D. G., True L. D., Okamoto A., Pomeroy S. L., Singer S., Golub T. R., Lander E. S., Getz G., Sellers W. R., and Meyerson M. (2010) The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

