Gut biogeography of the bacterial microbiota
- PMID: 26499895
- PMCID: PMC4837114
- DOI: 10.1038/nrmicro3552
Gut biogeography of the bacterial microbiota
Abstract
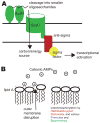
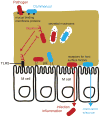
Animals assemble and maintain a diverse but host-specific gut microbial community. In addition to characteristic microbial compositions along the longitudinal axis of the intestines, discrete bacterial communities form in microhabitats, such as the gut lumen, colonic mucus layers and colonic crypts. In this Review, we examine how the spatial distribution of symbiotic bacteria among physical niches in the gut affects the development and maintenance of a resilient microbial ecosystem. We consider novel hypotheses for how nutrient selection, immune activation and other mechanisms control the biogeography of bacteria in the gut, and we discuss the relevance of this spatial heterogeneity to health and disease.
Figures




References
-
- Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol. 2011;9:356–368. - PubMed
-
- Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–339. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

