The Differences Between Cis- and Trans-Gene Inactivation Caused by Heterochromatin in Drosophila
- PMID: 26500261
- PMCID: PMC4701106
- DOI: 10.1534/genetics.115.181693
The Differences Between Cis- and Trans-Gene Inactivation Caused by Heterochromatin in Drosophila
Abstract
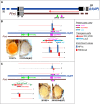
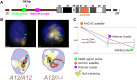
Position-effect variegation (PEV) is the epigenetic disruption of gene expression near the de novo-formed euchromatin-heterochromatin border. Heterochromatic cis-inactivation may be accompanied by the trans-inactivation of genes on a normal homologous chromosome in trans-heterozygous combination with a PEV-inducing rearrangement. We characterize a new genetic system, inversion In(2)A4, demonstrating cis-acting PEV as well as trans-inactivation of the reporter transgenes on the homologous nonrearranged chromosome. The cis-effect of heterochromatin in the inversion results not only in repression but also in activation of genes, and it varies at different developmental stages. While cis-actions affect only a few juxtaposed genes, trans-inactivation is observed in a 500-kb region and demonstrates а nonuniform pattern of repression with intermingled regions where no transgene repression occurs. There is no repression around the histone gene cluster and in some other euchromatic sites. trans-Inactivation is accompanied by dragging of euchromatic regions into the heterochromatic compartment, but the histone gene cluster, located in the middle of the trans-inactivated region, was shown to be evicted from the heterochromatin. We demonstrate that trans-inactivation is followed by de novo HP1a accumulation in the affected transgene; trans-inactivation is specifically favored by the chromatin remodeler SAYP and prevented by Argonaute AGO2.
Keywords: Drosophila; PEV; heterochromatin; nuclear compartmentalization; trans-inactivation.
Copyright © 2016 by the Genetics Society of America.
Figures




References
-
- Abramov Y. A., Kibanov M. V., Gvozdev V. A., Lavrov S. A., 2011. Genetic and molecular analysis of gene trans-inactivation caused by homologous eu-heterochromatic chromosome rearrangement in Drosophila melanogaster. Dokl. Biochem. Biophys. 437: 72–76. - PubMed
-
- Cryderman D. E., Grade S. K., Li Y., Fanti L., Pimpinelli S., et al. , 2005. Role of Drosophila HP1 in euchromatic gene expression. Dev. Dyn. 232: 767–774. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

