Human microbiomes and their roles in dysbiosis, common diseases, and novel therapeutic approaches
- PMID: 26500616
- PMCID: PMC4594012
- DOI: 10.3389/fmicb.2015.01050
Human microbiomes and their roles in dysbiosis, common diseases, and novel therapeutic approaches
Abstract
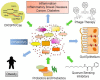
The human body is the residence of a large number of commensal (non-pathogenic) and pathogenic microbial species that have co-evolved with the human genome, adaptive immune system, and diet. With recent advances in DNA-based technologies, we initiated the exploration of bacterial gene functions and their role in human health. The main goal of the human microbiome project is to characterize the abundance, diversity and functionality of the genes present in all microorganisms that permanently live in different sites of the human body. The gut microbiota expresses over 3.3 million bacterial genes, while the human genome expresses only 20 thousand genes. Microbe gene-products exert pivotal functions via the regulation of food digestion and immune system development. Studies are confirming that manipulation of non-pathogenic bacterial strains in the host can stimulate the recovery of the immune response to pathogenic bacteria causing diseases. Different approaches, including the use of nutraceutics (prebiotics and probiotics) as well as phages engineered with CRISPR/Cas systems and quorum sensing systems have been developed as new therapies for controlling dysbiosis (alterations in microbial community) and common diseases (e.g., diabetes and obesity). The designing and production of pharmaceuticals based on our own body's microbiome is an emerging field and is rapidly growing to be fully explored in the near future. This review provides an outlook on recent findings on the human microbiomes, their impact on health and diseases, and on the development of targeted therapies.
Keywords: CRISPR/Cas system; metagenomics; microbiome; phage therapy; pharmacomicrobiomics; quorum sensing.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

