Diverse electron sources support denitrification under hypoxia in the obligate methanotroph Methylomicrobium album strain BG8
- PMID: 26500622
- PMCID: PMC4594100
- DOI: 10.3389/fmicb.2015.01072
Diverse electron sources support denitrification under hypoxia in the obligate methanotroph Methylomicrobium album strain BG8
Abstract
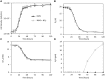
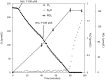
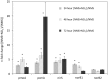
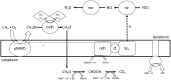
Aerobic methane-oxidizing bacteria (MOB) are a diverse group of microorganisms that are ubiquitous in natural environments. Along with anaerobic MOB and archaea, aerobic methanotrophs are critical for attenuating emission of methane to the atmosphere. Clearly, nitrogen availability in the form of ammonium and nitrite have strong effects on methanotrophic activity and their natural community structures. Previous findings show that nitrite amendment inhibits the activity of some cultivated methanotrophs; however, the physiological pathways that allow some strains to transform nitrite, expression of gene inventories, as well as the electron sources that support this activity remain largely uncharacterized. Here we show that Methylomicrobium album strain BG8 utilizes methane, methanol, formaldehyde, formate, ethane, ethanol, and ammonia to support denitrification activity under hypoxia only in the presence of nitrite. We also demonstrate that transcript abundance of putative denitrification genes, nirS and one of two norB genes, increased in response to nitrite. Furthermore, we found that transcript abundance of pxmA, encoding the alpha subunit of a putative copper-containing monooxygenase, increased in response to both nitrite and hypoxia. Our results suggest that expression of denitrification genes, found widely within genomes of aerobic methanotrophs, allow the coupling of substrate oxidation to the reduction of nitrogen oxide terminal electron acceptors under oxygen limitation. The present study expands current knowledge of the metabolic flexibility of methanotrophs by revealing that a diverse array of electron donors support nitrite reduction to nitrous oxide under hypoxia.
Keywords: Methylomicrobium album BG8; denitrification; hypoxia; methane monooxygenase; methanotroph; nitrite reduction; nitrous oxide.
Figures


References
-
- Bodelier P. L. E., Steenbergh A. K. (2014). Interactions between methane and the nitrogen cycle in light of climate change. Curr. Opin. Environ. Sustain. 9–10, 26–36. 10.1016/j.cosust.2014.07.004 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

