Genome-wide identification of microRNAs regulating cholesterol and triglyceride homeostasis
- PMID: 26501192
- PMCID: PMC4993048
- DOI: 10.1038/nm.3980
Genome-wide identification of microRNAs regulating cholesterol and triglyceride homeostasis
Abstract
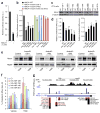
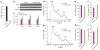
Genome-wide association studies (GWASs) have linked genes to various pathological traits. However, the potential contribution of regulatory noncoding RNAs, such as microRNAs (miRNAs), to a genetic predisposition to pathological conditions has remained unclear. We leveraged GWAS meta-analysis data from >188,000 individuals to identify 69 miRNAs in physical proximity to single-nucleotide polymorphisms (SNPs) associated with abnormal levels of circulating lipids. Several of these miRNAs (miR-128-1, miR-148a, miR-130b, and miR-301b) control the expression of key proteins involved in cholesterol-lipoprotein trafficking, such as the low-density lipoprotein (LDL) receptor (LDLR) and the ATP-binding cassette A1 (ABCA1) cholesterol transporter. Consistent with human liver expression data and genetic links to abnormal blood lipid levels, overexpression and antisense targeting of miR-128-1 or miR-148a in high-fat diet-fed C57BL/6J and Apoe-null mice resulted in altered hepatic expression of proteins involved in lipid trafficking and metabolism, and in modulated levels of circulating lipoprotein-cholesterol and triglycerides. Taken together, these findings support the notion that altered expression of miRNAs may contribute to abnormal blood lipid levels, predisposing individuals to human cardiometabolic disorders.
Figures

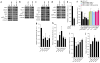
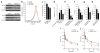
References
Publication types
MeSH terms
Substances
Grants and funding
- P30 DK040561/DK/NIDDK NIH HHS/United States
- R21DK084459/DK/NIDDK NIH HHS/United States
- P30 NS072030/NS/NINDS NIH HHS/United States
- K24 DK078772/DK/NIDDK NIH HHS/United States
- R01 HL107953/HL/NHLBI NIH HHS/United States
- R01 DK056626/DK/NIDDK NIH HHS/United States
- R01 HL049094/HL/NHLBI NIH HHS/United States
- R01HL106063/HL/NHLBI NIH HHS/United States
- R01DK094184/DK/NIDDK NIH HHS/United States
- K24DK078772/DK/NIDDK NIH HHS/United States
- R37 DK048873/DK/NIDDK NIH HHS/United States
- P30 DK034854/DK/NIDDK NIH HHS/United States
- R01 DK094184/DK/NIDDK NIH HHS/United States
- K99 HG008179/HG/NHGRI NIH HHS/United States
- R01HL107953/HL/NHLBI NIH HHS/United States
- R21 DK084459/DK/NIDDK NIH HHS/United States
- R01 HL106063/HL/NHLBI NIH HHS/United States
- R37DK048873/DK/NIDDK NIH HHS/United States
- R01DK056626/DK/NIDDK NIH HHS/United States
- R01HL49094/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

