Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements
- PMID: 26501517
- PMCID: PMC4666778
- DOI: 10.1038/nmeth.3630
Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements
Abstract
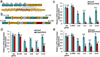
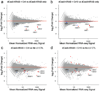
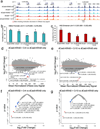
Epigenome editing with the CRISPR (clustered, regularly interspaced, short palindromic repeats)-Cas9 platform is a promising technology for modulating gene expression to direct cell phenotype and to dissect the causal epigenetic mechanisms of gene regulation. Fusions of nuclease-inactive dCas9 to the Krüppel-associated box (KRAB) repressor (dCas9-KRAB) can silence target gene expression, but the genome-wide specificity and the extent of heterochromatin formation catalyzed by dCas9-KRAB are not known. We targeted dCas9-KRAB to the HS2 enhancer, a distal regulatory element that orchestrates the expression of multiple globin genes, and observed highly specific induction of H3K9 trimethylation (H3K9me3) at the enhancer and decreased chromatin accessibility of both the enhancer and its promoter targets. Targeted epigenetic modification of HS2 silenced the expression of multiple globin genes, with minimal off-target changes in global gene expression. These results demonstrate that repression mediated by dCas9-KRAB is sufficiently specific to disrupt the activity of individual enhancers via local modification of the epigenome.
Figures


References
-
- Snowden AW, Gregory PD, Case CC, Pabo CO. Gene-specific targeting of H3K9 methylation is sufficient for initiating repression in vivo. Curr Biol. 2002;12:2159–2166. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

