A comparison of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs
- PMID: 26501951
- PMCID: PMC4847940
- DOI: 10.1038/nbt.3388
A comparison of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs
Abstract
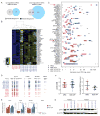
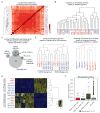
The equivalence of human induced pluripotent stem cells (hiPSCs) and human embryonic stem cells (hESCs) remains controversial. Here we use genetically matched hESC and hiPSC lines to assess the contribution of cellular origin (hESC vs. hiPSC), the Sendai virus (SeV) reprogramming method and genetic background to transcriptional and DNA methylation patterns while controlling for cell line clonality and sex. We find that transcriptional and epigenetic variation originating from genetic background dominates over variation due to cellular origin or SeV infection. Moreover, the 49 differentially expressed genes we detect between genetically matched hESCs and hiPSCs neither predict functional outcome nor distinguish an independently derived, larger set of unmatched hESC and hiPSC lines. We conclude that hESCs and hiPSCs are molecularly and functionally equivalent and cannot be distinguished by a consistent gene expression signature. Our data further imply that genetic background variation is a major confounding factor for transcriptional and epigenetic comparisons of pluripotent cell lines, explaining some of the previously observed differences between genetically unmatched hESCs and hiPSCs.
Figures



Comment in
-
Putting induced pluripotent stem cells to the test.Nat Biotechnol. 2015 Nov;33(11):1145-6. doi: 10.1038/nbt.3401. Nat Biotechnol. 2015. PMID: 26544143 No abstract available.
References
-
- Takahashi KK, Yamanaka SS. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. - PubMed
-
- Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. - PubMed
-
- Yu J, et al. Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. Science. 2007;318:1917–1920. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

