Integrated analysis of genome-wide DNA methylation, gene expression and protein expression profiles in molecular subtypes of WHO II-IV gliomas
- PMID: 26502731
- PMCID: PMC4620600
- DOI: 10.1186/s13046-015-0249-z
Integrated analysis of genome-wide DNA methylation, gene expression and protein expression profiles in molecular subtypes of WHO II-IV gliomas
Abstract
Background: Glioma is the most common malignant primary brain tumor among adults, among which glioblastoma (GBM) exhibits the highest malignancy. Despite current standard chemoradiation, glioma is still invariably fatal. A further insight into the molecular background of glioma is required to improve patient outcomes.
Method: Previous studies evaluated molecular genetic differences through comparing different grades of glioma. Here, we integrated DNA methylation, RNA sequencing and protein expression data sets of WHO grade II to IV gliomas, to screen for dysregulated genes in subtypes during malignant progression of glioma.
Results: We propose a list of universal genes (UG) as novel glioma biomarkers: 977 up-regulated genes and 114 down-regulated genes, who involved in cell cycle, Wnt receptor signaling pathway and fatty acid metabolic process. Poorer survival was associated significantly with the high expression of 977 up-regulated genes and low expression of 114 down-regulated in UG (P <0.001).
Conclusion: To our knowledge, this was the first study that focused on subtypes to detect dysregulated genes that could contribute to malignant progression. Furthermore, the differentially expressed genes profile may lead to the identification of new therapeutic targets for glioma patients.
Figures
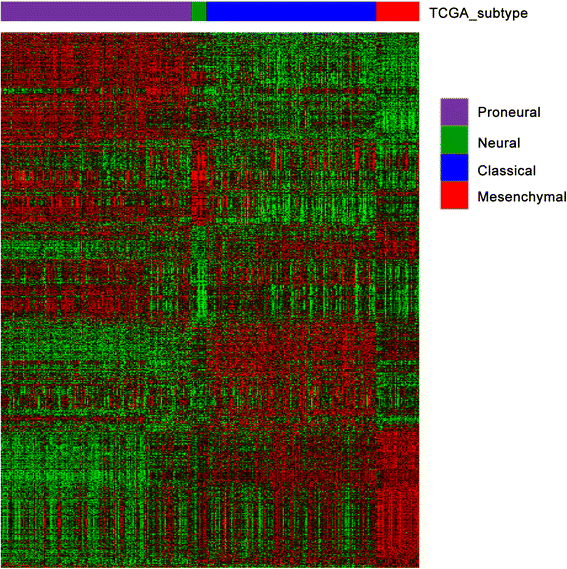
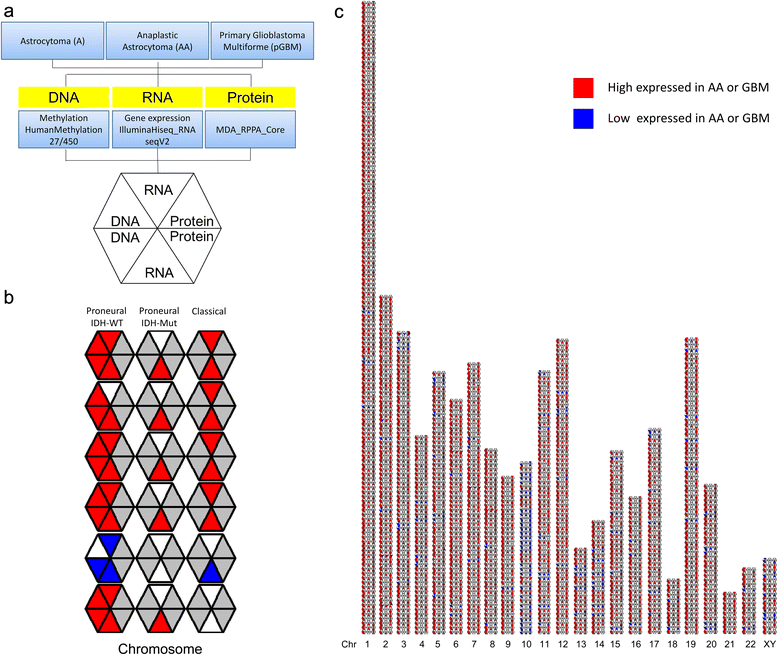


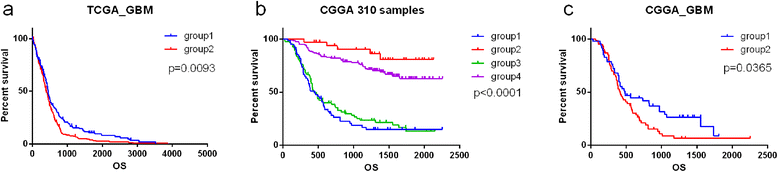
References
-
- Cai J, Yang P, Zhang C, Zhang W, Liu Y, Bao Z, et al. ATRX mRNA expression combined with IDH1/2 mutational status and Ki-67 expression refines the molecular classification of astrocytic tumors: evidence from the whole transcriptome sequencing of 169 samples samples. Oncotarget. 2014;15(9):2551–61. doi: 10.18632/oncotarget.1838. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

