Evaluation of TCR Gene Editing Achieved by TALENs, CRISPR/Cas9, and megaTAL Nucleases
- PMID: 26502778
- PMCID: PMC4786913
- DOI: 10.1038/mt.2015.197
Evaluation of TCR Gene Editing Achieved by TALENs, CRISPR/Cas9, and megaTAL Nucleases
Abstract
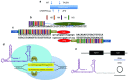
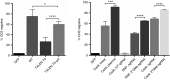
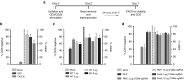
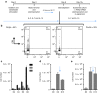
Present adoptive immunotherapy strategies are based on the re-targeting of autologous T-cells to recognize tumor antigens. As T-cell properties may vary significantly between patients, this approach can result in significant variability in cell potency that may affect therapeutic outcome. More consistent results could be achieved by generating allogeneic cells from healthy donors. An impediment to such an approach is the endogenous T-cell receptors present on T-cells, which have the potential to direct dangerous off-tumor antihost reactivity. To address these limitations, we assessed the ability of three different TCR-α-targeted nucleases to disrupt T-cell receptor expression in primary human T-cells. We optimized the conditions for the delivery of each reagent and assessed off-target cleavage. The megaTAL and CRISPR/Cas9 reagents exhibited the highest disruption efficiency combined with low levels of toxicity and off-target cleavage, and we used them for a translatable manufacturing process to produce safe cellular substrates for next-generation immunotherapies.
Figures

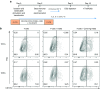
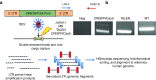
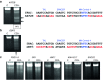
References
-
- Berdien, B, Mock, U, Atanackovic, D and Fehse, B (2014). TALEN-mediated editing of endogenous T-cell receptors facilitates efficient reprogramming of T lymphocytes by lentiviral gene transfer. Gene Ther 21: 539–548. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

