CMT2D neuropathy is linked to the neomorphic binding activity of glycyl-tRNA synthetase
- PMID: 26503042
- PMCID: PMC4754353
- DOI: 10.1038/nature15510
CMT2D neuropathy is linked to the neomorphic binding activity of glycyl-tRNA synthetase
Erratum in
-
Corrigendum: CMT2D neuropathy is linked to the neomorphic binding activity of glycyl-tRNA synthetase.Nature. 2016 Apr 21;532(7599):402. doi: 10.1038/nature16499. Epub 2016 Jan 20. Nature. 2016. PMID: 26789244 No abstract available.
Abstract
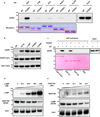
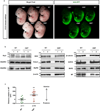
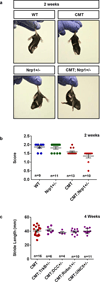
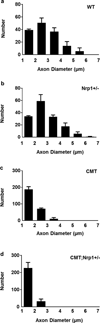
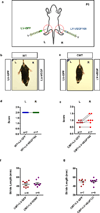
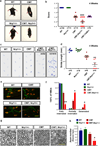
Selective neuronal loss is a hallmark of neurodegenerative diseases, which, counterintuitively, are often caused by mutations in widely expressed genes. Charcot-Marie-Tooth (CMT) diseases are the most common hereditary peripheral neuropathies, for which there are no effective therapies. A subtype of these diseases--CMT type 2D (CMT2D)--is caused by dominant mutations in GARS, encoding the ubiquitously expressed enzyme glycyl-transfer RNA (tRNA) synthetase (GlyRS). Despite the broad requirement of GlyRS for protein biosynthesis in all cells, mutations in this gene cause a selective degeneration of peripheral axons, leading to deficits in distal motor function. How mutations in GlyRS (GlyRS(CMT2D)) are linked to motor neuron vulnerability has remained elusive. Here we report that GlyRS(CMT2D) acquires a neomorphic binding activity that directly antagonizes an essential signalling pathway for motor neuron survival. We find that CMT2D mutations alter the conformation of GlyRS, enabling GlyRS(CMT2D) to bind the neuropilin 1 (Nrp1) receptor. This aberrant interaction competitively interferes with the binding of the cognate ligand vascular endothelial growth factor (VEGF) to Nrp1. Genetic reduction of Nrp1 in mice worsens CMT2D symptoms, whereas enhanced expression of VEGF improves motor function. These findings link the selective pathology of CMT2D to the neomorphic binding activity of GlyRS(CMT2D) that antagonizes the VEGF-Nrp1 interaction, and indicate that the VEGF-Nrp1 signalling axis is an actionable target for treating CMT2D.
Figures








References
-
- Saxena S, Caroni P. Selective neuronal vulnerability in neurodegenerative diseases: from stressor thresholds to degeneration. Neuron. 2011;71:35–48. - PubMed
-
- Skre H. Genetic and clinical aspects of Charcot-Marie-Tooth's disease. Clin Genet. 1974;6:98–118. - PubMed
-
- Wakasugi K, Schimmel P. Two distinct cytokines released from a human aminoacyl-tRNA synthetase. Science. 1999;284:147–151. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

