The strigolactone biosynthesis gene DWARF27 is co-opted in rhizobium symbiosis
- PMID: 26503135
- PMCID: PMC4624177
- DOI: 10.1186/s12870-015-0651-x
The strigolactone biosynthesis gene DWARF27 is co-opted in rhizobium symbiosis
Abstract
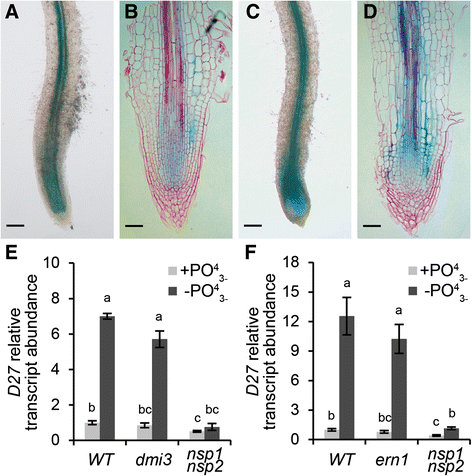
Background: Strigolactones are a class of plant hormones whose biosynthesis is activated in response to phosphate starvation. This involves several enzymes, including the carotenoid cleavage dioxygenases 7 (CCD7) and CCD8 and the carotenoid isomerase DWARF27 (D27). D27 expression is known to be responsive to phosphate starvation. In Medicago truncatula and rice (Oryza sativa) this transcriptional response requires the GRAS-type proteins NSP1 and NSP2; both proteins are essential for rhizobium induced root nodule formation in legumes. In line with this, we questioned whether MtNSP1-MtNSP2 dependent MtD27 regulation is co-opted in rhizobium symbiosis.
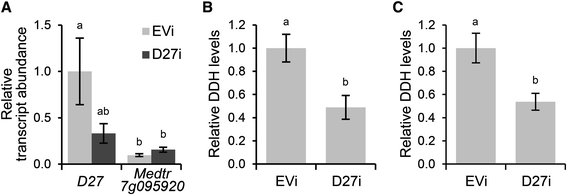
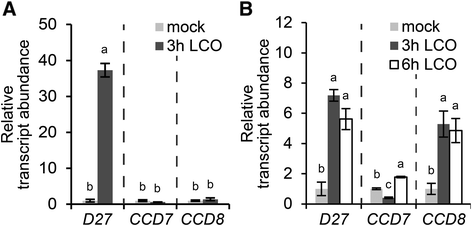
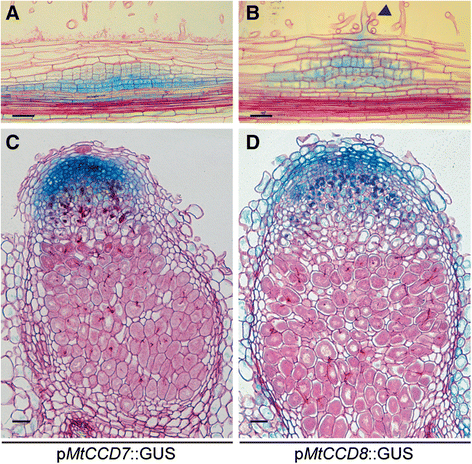
Results: We provide evidence that MtD27 is involved in strigolactone biosynthesis in M. truncatula roots upon phosphate stress. Spatiotemporal expression studies revealed that this gene is also highly expressed in nodule primordia and subsequently becomes restricted to the meristem and distal infection zone of a mature nodules. A similar expression pattern was found for MtCCD7 and MtCCD8. Rhizobium lipo-chitooligosaccharide (LCO) application experiments revealed that of these genes MtD27 is most responsive in an MtNSP1 and MtNSP2 dependent manner. Symbiotic expression of MtD27 requires components of the symbiosis signaling pathway; including MtDMI1, MtDMI2, MtDMI3/MtCCaMK and in part MtERN1. This in contrast to MtD27 expression upon phosphate starvation, which only requires MtNSP1 and MtNSP2.
Conclusion: Our data show that the phosphate-starvation responsive strigolactone biosynthesis gene MtD27 is also rapidly induced by rhizobium LCO signals in an MtNSP1 and MtNSP2-dependent manner. Additionally, we show that MtD27 is co-expressed with MtCCD7 and MtCCD8 in nodule primordia and in the infection zone of mature nodules.
Figures





Similar articles
-
Rhizobium Lipo-chitooligosaccharide Signaling Triggers Accumulation of Cytokinins in Medicago truncatula Roots.Mol Plant. 2015 Aug;8(8):1213-26. doi: 10.1016/j.molp.2015.03.010. Epub 2015 Mar 21. Mol Plant. 2015. PMID: 25804975
-
Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2.Plant Cell. 2011 Oct;23(10):3853-65. doi: 10.1105/tpc.111.089771. Epub 2011 Oct 28. Plant Cell. 2011. PMID: 22039214 Free PMC article.
-
Deep Sequencing of the Medicago truncatula Root Transcriptome Reveals a Massive and Early Interaction between Nodulation Factor and Ethylene Signals.Plant Physiol. 2015 Sep;169(1):233-65. doi: 10.1104/pp.15.00350. Epub 2015 Jul 14. Plant Physiol. 2015. PMID: 26175514 Free PMC article.
-
Strigolactone signaling in root development and phosphate starvation.Plant Signal Behav. 2015;10(7):e1045174. doi: 10.1080/15592324.2015.1045174. Plant Signal Behav. 2015. PMID: 26251884 Free PMC article. Review.
-
From carotenoids to strigolactones.J Exp Bot. 2018 Apr 23;69(9):2189-2204. doi: 10.1093/jxb/erx476. J Exp Bot. 2018. PMID: 29253188 Review.
Cited by
-
Chemotactic Host-Finding Strategies of Plant Endoparasites and Endophytes.Front Plant Sci. 2020 Jul 31;11:1167. doi: 10.3389/fpls.2020.01167. eCollection 2020. Front Plant Sci. 2020. PMID: 32849722 Free PMC article. Review.
-
Nutrient regulation of lipochitooligosaccharide recognition in plants via NSP1 and NSP2.Nat Commun. 2022 Oct 28;13(1):6421. doi: 10.1038/s41467-022-33908-3. Nat Commun. 2022. PMID: 36307431 Free PMC article.
-
Strigolactone Levels in Dicot Roots Are Determined by an Ancestral Symbiosis-Regulated Clade of the PHYTOENE SYNTHASE Gene Family.Front Plant Sci. 2018 Mar 1;9:255. doi: 10.3389/fpls.2018.00255. eCollection 2018. Front Plant Sci. 2018. PMID: 29545815 Free PMC article.
-
The Role of Strigolactone in the Cross-Talk Between Arabidopsis thaliana and the Endophytic Fungus Mucor sp.Front Microbiol. 2018 Mar 19;9:441. doi: 10.3389/fmicb.2018.00441. eCollection 2018. Front Microbiol. 2018. PMID: 29615990 Free PMC article.
-
Signaling in Legume-Rhizobia Symbiosis.Int J Mol Sci. 2023 Dec 12;24(24):17397. doi: 10.3390/ijms242417397. Int J Mol Sci. 2023. PMID: 38139226 Free PMC article. Review.
References
-
- Soto MJ, Fernández-Aparicio M, Castellanos-Morales V, García-Garrido JM, Ocampo JA, Delgado MJ, et al. First indications for the involvement of strigolactones on nodule formation in alfalfa (Medicago sativa) Soil Biol Biochem. 2010;42:383–385. doi: 10.1016/j.soilbio.2009.11.007. - DOI
-
- Liu J, Novero M, Charnikhova T, Ferrandino A, Schubert A, Ruyter-Spira C, et al. Carotenoid cleavage dioxygenase 7 modulates plant growth, reproduction, senescence, and determinate nodulation in the model legume Lotus japonicus. J Exp Bot. 2013;64:1967–1981. doi: 10.1093/jxb/ert056. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

