Whole Exome Sequencing Identifies Frequent Somatic Mutations in Cell-Cell Adhesion Genes in Chinese Patients with Lung Squamous Cell Carcinoma
- PMID: 26503331
- PMCID: PMC4621504
- DOI: 10.1038/srep14237
Whole Exome Sequencing Identifies Frequent Somatic Mutations in Cell-Cell Adhesion Genes in Chinese Patients with Lung Squamous Cell Carcinoma
Abstract
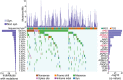
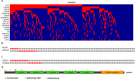
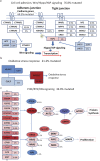
Lung squamous cell carcinoma (SQCC) accounts for about 30% of all lung cancer cases. Understanding of mutational landscape for this subtype of lung cancer in Chinese patients is currently limited. We performed whole exome sequencing in samples from 100 patients with lung SQCCs to search for somatic mutations and the subsequent target capture sequencing in another 98 samples for validation. We identified 20 significantly mutated genes, including TP53, CDH10, NFE2L2 and PTEN. Pathways with frequently mutated genes included those of cell-cell adhesion/Wnt/Hippo in 76%, oxidative stress response in 21%, and phosphatidylinositol-3-OH kinase in 36% of the tested tumor samples. Mutations of Chromatin regulatory factor genes were identified at a lower frequency. In functional assays, we observed that knockdown of CDH10 promoted cell proliferation, soft-agar colony formation, cell migration and cell invasion, and overexpression of CDH10 inhibited cell proliferation. This mutational landscape of lung SQCC in Chinese patients improves our current understanding of lung carcinogenesis, early diagnosis and personalized therapy.
Figures

References
-
- Jemal A. et al. Global cancer statistics. CA Cancer J Clin 61, 69–90 (2011). - PubMed
-
- Jemal A., Siegel R., Xu J. & Ward E. Cancer statistics, 2010. CA Cancer J Clin 60, 277–300 (2010). - PubMed
-
- Maemondo M. et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 362, 2380–2388 (2010). - PubMed
-
- Gao W. M. et al. Comparison of p53 mutations between adenocarcinoma and squamous cell carcinoma of the lung: unique spectra involving G to A transitions and G to T transversions in both histologic types. Lung Cancer 40, 141–150 (2003). - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

