Polymorphisms involving gain or loss of CpG sites are significantly enriched in trait-associated SNPs
- PMID: 26503467
- PMCID: PMC4741875
- DOI: 10.18632/oncotarget.5650
Polymorphisms involving gain or loss of CpG sites are significantly enriched in trait-associated SNPs
Abstract
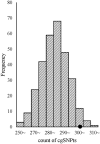
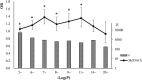
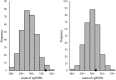
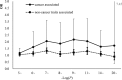
Some single nucleotide polymorphisms (SNPs) influence the existence of CpG sites, the basis of DNA modification such as methylation and hydroxymethylation. These polymorphisms can lead to gain or loss of CpG sites and were defined as CpG site related SNPs (cgSNPs) in this study. The cgSNPs change DNA sequence and might potentially affect DNA modification such as methylation. However, the functional consequence of cgSNPs is poorly understood. We observed that a considerable proportion (23.0%) of common variants were cgSNPs in human genome. Mutations involving loss of CpG sites were associated with reduced levels of methylation (~20.2%) using The Cancer Genome Atlas (TCGA) data. Using public databases (SCAN and seeQTL) of expression quantitative trait loci (eQTLs), we found that the cgSNPs were significantly enriched in eQTLs via logistic regression and simulation test. Furthermore, we observed that cgSNPs were more likely to be trait-associated loci especially cancers using a catalog of published genome-wide association studies (GWAS) recorded by National Human Genome Research Institute (NHGRI). Our results indicated that cgSNP might be meaningful as annotation either in SNP functional prediction or in screening for trait-associated SNPs.
Keywords: CpG site; DNA methylation; cancer; epigenetic; single nucleotide polymorphism (SNP).
Conflict of interest statement
No potential conflicts of interest was disclosed.
Figures


References
-
- Bird A. DNA methylation patterns and epigenetic memory. Genes & development. 2002;16:6–21. - PubMed
-
- Jabbari K, Bernardi G. Cytosine methylation and CpG, TpG (CpA) and TpA frequencies. Gene. 2004;333:143–149. - PubMed
-
- Razin A, Riggs AD. DNA methylation and gene function. Science. 1980;210:604–610. - PubMed
-
- Bergman Y, Cedar H. DNA methylation dynamics in health and disease. Nature structural & molecular biology. 2013;20:274–281. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

