Multiplexed, Proteome-Wide Protein Expression Profiling: Yeast Deubiquitylating Enzyme Knockout Strains
- PMID: 26503604
- PMCID: PMC4802358
- DOI: 10.1021/acs.jproteome.5b00802
Multiplexed, Proteome-Wide Protein Expression Profiling: Yeast Deubiquitylating Enzyme Knockout Strains
Abstract
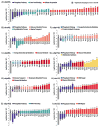
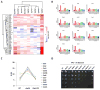
Characterizing a protein's function often requires a description of the cellular state in its absence. Multiplexing in mass spectrometry-based proteomics has now achieved the ability to globally measure protein expression levels in yeast from 10 cell states simultaneously. We applied this approach to quantify expression differences in wild type and nine deubiquitylating enzyme (DUB) knockout strains with the goal of creating "information networks" that might provide deeper, mechanistic insights into a protein's biological role. In total, more than 3700 proteins were quantified with high reproducibility across three biological replicates (30 samples in all). DUB mutants demonstrated different proteomics profiles, consistent with distinct roles for each family member. These included differences in total ubiquitin levels and specific chain linkages. Moreover, specific expression changes suggested novel functions for several DUB family members. For instance, the ubp3Δ mutant showed large expression changes for members of the cytochrome C oxidase complex, consistent with a role for Ubp3 in mitochondrial regulation. Several DUBs also showed broad expression changes for phosphate transporters as well as other components of the inorganic phosphate signaling pathway, suggesting a role for these DUBs in regulating phosphate metabolism. These data highlight the potential of multiplexed proteome-wide analyses for biological investigation and provide a framework for further study of the DUB family. Our methods are readily applicable to the entire collection of yeast deletion mutants and may help facilitate systematic analysis of yeast and other organisms.
Keywords: COX complex; Orbitrap Fusion; TMT; UBP3; deubiquitinases; high-throughput proteomics; inorganic phosphate pathway; isobaric labeling; quantitative proteomics; ubiquitin.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




References
-
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M’Rabet N, Menard P, Mittmann M, Pai C, Rebischung C, Revuelta JL, Riles L, Roberts CJ, Ross-MacDonald P, Scherens B, Snyder M, Sookhai-Mahadeo S, Storms RK, Veronneau S, Voet M, Volckaert G, Ward TR, Wysocki R, Yen GS, Yu K, Zimmermann K, Philippsen P, Johnston M, Davis RW. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285(5429):901–6. - PubMed
-
- Steinmetz LM, Scharfe C, Deutschbauer AM, Mokranjac D, Herman ZS, Jones T, Chu AM, Giaever G, Prokisch H, Oefner PJ, Davis RW. Systematic screen for human disease genes in yeast. Nat Genet. 2002;31(4):400–4. - PubMed
-
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Guldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kotter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang CY, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418(6896):387–91. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

