Why significant variables aren't automatically good predictors
- PMID: 26504198
- PMCID: PMC4653162
- DOI: 10.1073/pnas.1518285112
Why significant variables aren't automatically good predictors
Abstract
Thus far, genome-wide association studies (GWAS) have been disappointing in the inability of investigators to use the results of identified, statistically significant variants in complex diseases to make predictions useful for personalized medicine. Why are significant variables not leading to good prediction of outcomes? We point out that this problem is prevalent in simple as well as complex data, in the sciences as well as the social sciences. We offer a brief explanation and some statistical insights on why higher significance cannot automatically imply stronger predictivity and illustrate through simulations and a real breast cancer example. We also demonstrate that highly predictive variables do not necessarily appear as highly significant, thus evading the researcher using significance-based methods. We point out that what makes variables good for prediction versus significance depends on different properties of the underlying distributions. If prediction is the goal, we must lay aside significance as the only selection standard. We suggest that progress in prediction requires efforts toward a new research agenda of searching for a novel criterion to retrieve highly predictive variables rather than highly significant variables. We offer an alternative approach that was not designed for significance, the partition retention method, which was very effective predicting on a long-studied breast cancer data set, by reducing the classification error rate from 30% to 8%.
Keywords: high-dimensional data; prediction; statistical significance; variable selection classification.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
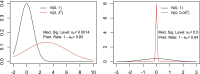



References
-
- Anonymous Predicting the influence of common variants. Nat Genet. 2013;45(4):339. - PubMed
-
- de los Campos G, Gianola D, Allison DB. Predicting genetic predisposition in humans: The promise of whole-genome markers. Nat Rev Genet. 2010;11(12):880–886. - PubMed
-
- Janssens AC, van Duijn CM. Genome-based prediction of common diseases: advances and prospects. Hum Mol Genet. 2008;17(R2):R166–R173. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

