Nanoscale spatial organization of the HoxD gene cluster in distinct transcriptional states
- PMID: 26504220
- PMCID: PMC4653165
- DOI: 10.1073/pnas.1517972112
Nanoscale spatial organization of the HoxD gene cluster in distinct transcriptional states
Abstract
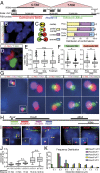
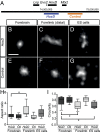
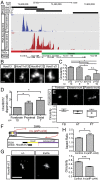
Chromatin condensation plays an important role in the regulation of gene expression. Recently, it was shown that the transcriptional activation of Hoxd genes during vertebrate digit development involves modifications in 3D interactions within and around the HoxD gene cluster. This reorganization follows a global transition from one set of regulatory contacts to another, between two topologically associating domains (TADs) located on either side of the HoxD locus. Here, we use 3D DNA FISH to assess the spatial organization of chromatin at and around the HoxD gene cluster and report that although the two TADs are tightly associated, they appear as spatially distinct units. We measured the relative position of genes within the cluster and found that they segregate over long distances, suggesting that a physical elongation of the HoxD cluster can occur. We analyzed this possibility by super-resolution imaging (STORM) and found that tissues with distinct transcriptional activity exhibit differing degrees of elongation. We also observed that the morphological change of the HoxD cluster in developing digits is associated with its position at the boundary between the two TADs. Such variations in the fine-scale architecture of the gene cluster suggest causal links among its spatial configuration, transcriptional activation, and the flanking chromatin context.
Keywords: DNA FISH; HoxD; limb development; super-resolution microscopy; topologically associating domains.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Bickmore WA, van Steensel B. Genome architecture: Domain organization of interphase chromosomes. Cell. 2013;152(6):1270–1284. - PubMed
-
- de Laat W, Duboule D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature. 2013;502(7472):499–506. - PubMed
-
- Chambeyron S, Bickmore WA. Does looping and clustering in the nucleus regulate gene expression? Curr Opin Cell Biol. 2004;16(3):256–262. - PubMed
-
- Morey C, Da Silva NR, Perry P, Bickmore WA. Nuclear reorganisation and chromatin decondensation are conserved, but distinct, mechanisms linked to Hox gene activation. Development. 2007;134(5):909–919. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

