Tether mutations that restore function and suppress pleiotropic phenotypes of the C. elegans isp-1(qm150) Rieske iron-sulfur protein
- PMID: 26504246
- PMCID: PMC4653183
- DOI: 10.1073/pnas.1509416112
Tether mutations that restore function and suppress pleiotropic phenotypes of the C. elegans isp-1(qm150) Rieske iron-sulfur protein
Abstract
Mitochondria play an important role in numerous diseases as well as normative aging. Severe reduction in mitochondrial function contributes to childhood disorders such as Leigh Syndrome, whereas mild disruption can extend the lifespan of model organisms. The Caenorhabditis elegans isp-1 gene encodes the Rieske iron-sulfur protein subunit of cytochrome c oxidoreductase (complex III of the electron transport chain). The partial loss of function allele, isp-1(qm150), leads to several pleiotropic phenotypes. To better understand the molecular mechanisms of ISP-1 function, we sought to identify genetic suppressors of the delayed development of isp-1(qm150) animals. Here we report a series of intragenic suppressors, all located within a highly conserved six amino acid tether region of ISP-1. These intragenic mutations suppress all of the evaluated isp-1(qm150) phenotypes, including developmental rate, pharyngeal pumping rate, brood size, body movement, activation of the mitochondrial unfolded protein response reporter, CO2 production, mitochondrial oxidative phosphorylation, and lifespan extension. Furthermore, analogous mutations show a similar effect when engineered into the budding yeast Rieske iron-sulfur protein Rip1, revealing remarkable conservation of the structure-function relationship of these residues across highly divergent species. The focus on a single subunit as causal both in generation and in suppression of diverse pleiotropic phenotypes points to a common underlying molecular mechanism, for which we propose a "spring-loaded" model. These observations provide insights into how gating and control processes influence the function of ISP-1 in mediating pleiotropic phenotypes including developmental rate, movement, sensitivity to stress, and longevity.
Keywords: Rip1; aging; complex III; isp-1; mitochondrial iron–sulfur protein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

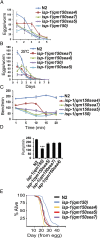

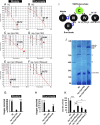


Comment in
-
New functional and biophysical insights into the mitochondrial Rieske iron-sulfur protein from genetic suppressor analysis in C. elegans.Worm. 2016 Apr 11;5(2):e1174803. doi: 10.1080/21624054.2016.1174803. eCollection 2016 Apr-Jun. Worm. 2016. PMID: 27383074 Free PMC article.
Similar articles
-
Two modes of mitochondrial dysfunction lead independently to lifespan extension in Caenorhabditis elegans.Aging Cell. 2010 Jun;9(3):433-47. doi: 10.1111/j.1474-9726.2010.00571.x. Epub 2010 Mar 19. Aging Cell. 2010. PMID: 20346072
-
New functional and biophysical insights into the mitochondrial Rieske iron-sulfur protein from genetic suppressor analysis in C. elegans.Worm. 2016 Apr 11;5(2):e1174803. doi: 10.1080/21624054.2016.1174803. eCollection 2016 Apr-Jun. Worm. 2016. PMID: 27383074 Free PMC article.
-
Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans.Dev Cell. 2001 Nov;1(5):633-44. doi: 10.1016/s1534-5807(01)00071-5. Dev Cell. 2001. PMID: 11709184
-
Respiratory complex III dysfunction in humans and the use of yeast as a model organism to study mitochondrial myopathy and associated diseases.Biochim Biophys Acta. 2013 Nov-Dec;1827(11-12):1346-61. doi: 10.1016/j.bbabio.2012.11.015. Epub 2012 Dec 5. Biochim Biophys Acta. 2013. PMID: 23220121 Review.
-
Mitochondrial respiration and reactive oxygen species in C. elegans.Exp Gerontol. 2006 Oct;41(10):957-67. doi: 10.1016/j.exger.2006.06.056. Epub 2006 Aug 21. Exp Gerontol. 2006. PMID: 16919906 Review.
Cited by
-
Hydrogels For Peptide Hormones Delivery: Therapeutic And Tissue Engineering Applications.Drug Des Devel Ther. 2019 Sep 26;13:3405-3418. doi: 10.2147/DDDT.S217211. eCollection 2019. Drug Des Devel Ther. 2019. PMID: 31579238 Free PMC article. Review.
-
The evolution of the human mitochondrial bc1 complex- adaptation for reduced rate of superoxide production?J Bioenerg Biomembr. 2023 Feb;55(1):15-31. doi: 10.1007/s10863-023-09957-8. Epub 2023 Feb 4. J Bioenerg Biomembr. 2023. PMID: 36737563
-
Mitonuclear Mismatch is Associated With Increased Male Frequency, Outcrossing, and Male Sperm Size in Experimentally-Evolved C. elegans.Front Genet. 2022 Mar 11;13:742272. doi: 10.3389/fgene.2022.742272. eCollection 2022. Front Genet. 2022. PMID: 35360860 Free PMC article.
-
Regulation of TERRA on telomeric and mitochondrial functions in IPF pathogenesis.BMC Pulm Med. 2017 Dec 2;17(1):163. doi: 10.1186/s12890-017-0516-1. BMC Pulm Med. 2017. PMID: 29197377 Free PMC article.
-
C. elegans electrotaxis behavior is modulated by heat shock response and unfolded protein response signaling pathways.Sci Rep. 2021 Feb 4;11(1):3115. doi: 10.1038/s41598-021-82466-z. Sci Rep. 2021. PMID: 33542359 Free PMC article.
References
-
- Wallace DC. Mitochondrial DNA mutations in disease and aging. Environ Mol Mutagen. 2010;51(5):440–450. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

