Transcriptional dynamics of the developing sweet cherry (Prunus avium L.) fruit: sequencing, annotation and expression profiling of exocarp-associated genes
- PMID: 26504533
- PMCID: PMC4591669
- DOI: 10.1038/hortres.2014.11
Transcriptional dynamics of the developing sweet cherry (Prunus avium L.) fruit: sequencing, annotation and expression profiling of exocarp-associated genes
Abstract
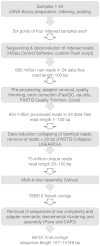
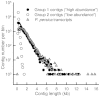
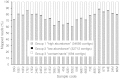
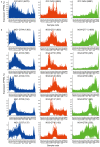
The exocarp, or skin, of fleshy fruit is a specialized tissue that protects the fruit, attracts seed dispersing fruit eaters, and has large economical relevance for fruit quality. Development of the exocarp involves regulated activities of many genes. This research analyzed global gene expression in the exocarp of developing sweet cherry (Prunus avium L., 'Regina'), a fruit crop species with little public genomic resources. A catalog of transcript models (contigs) representing expressed genes was constructed from de novo assembled short complementary DNA (cDNA) sequences generated from developing fruit between flowering and maturity at 14 time points. Expression levels in each sample were estimated for 34 695 contigs from numbers of reads mapping to each contig. Contigs were annotated functionally based on BLAST, gene ontology and InterProScan analyses. Coregulated genes were detected using partitional clustering of expression patterns. The results are discussed with emphasis on genes putatively involved in cuticle deposition, cell wall metabolism and sugar transport. The high temporal resolution of the expression patterns presented here reveals finely tuned developmental specialization of individual members of gene families. Moreover, the de novo assembled sweet cherry fruit transcriptome with 7760 full-length protein coding sequences and over 20 000 other, annotated cDNA sequences together with their developmental expression patterns is expected to accelerate molecular research on this important tree fruit crop.
Figures


References
-
- Esau K. Anatomy of Seed Plants. 2nd ed. New York: John Wiley & Sons, 1977.
-
- Baur P, Stulle K, Schönherr J, Uhlig B. [Absorption of radiation in the UV-B to blue light range by leave cuticles of selected crop plants].Gartenbauwissenschaft 1998; 63: 145–152. German
-
- Martin JT, Juniper BE. The Cuticles of Plants. Edinburgh: Edward Arnold Ltd, 1970.
-
- Kerstiens G. Plant Cuticles—An Integrated Functional Approach. Oxford: BIOS Scientific Publishers, 1996.
-
- Lalel HJD, Singh Z, Tan SC. Distribution of aroma volatile compounds in different parts of mango fruit. J Hort Sci Biotechnol 2003; 78: 131–138.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

