Mapping the Protein Fold Universe Using the CamTube Force Field in Molecular Dynamics Simulations
- PMID: 26505754
- PMCID: PMC4624779
- DOI: 10.1371/journal.pcbi.1004435
Mapping the Protein Fold Universe Using the CamTube Force Field in Molecular Dynamics Simulations
Abstract
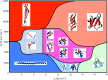
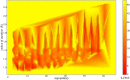
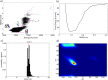
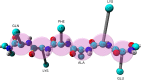
It has been recently shown that the coarse-graining of the structures of polypeptide chains as self-avoiding tubes can provide an effective representation of the conformational space of proteins. In order to fully exploit the opportunities offered by such a 'tube model' approach, we present here a strategy to combine it with molecular dynamics simulations. This strategy is based on the incorporation of the 'CamTube' force field into the Gromacs molecular dynamics package. By considering the case of a 60-residue polyvaline chain, we show that CamTube molecular dynamics simulations can comprehensively explore the conformational space of proteins. We obtain this result by a 20 μs metadynamics simulation of the polyvaline chain that recapitulates the currently known protein fold universe. We further show that, if residue-specific interaction potentials are added to the CamTube force field, it is possible to fold a protein into a topology close to that of its native state. These results illustrate how the CamTube force field can be used to explore efficiently the universe of protein folds with good accuracy and very limited computational cost.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Anfinsen C (1973) Principles that Govern the Folding of Protein Chains. Science 181: 223–230. - PubMed
-
- Bryngelson J, Onuchic J, Socci N, Wolynes P (1995) Funnels, Pathways and the Energy Landscape of Protein Folding: A Synthesis. Proteins 21: 167–195. - PubMed
-
- Dobson C (2003) Protein folding and misfolding. Nature 426: 884–890. - PubMed
-
- Onuchic J, Luthey-Schulten Z, Wolynes P (1997) Theory of protein folding: The energy landscape perspective. Annual Review of Physical Chemistry 48: 545–600. - PubMed
-
- Dill K, Chan HS (1997) From Levinthal to pathways to funnels. Nature Structural Biology 4: 10–19. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

