Novel RNA Duplex Locks HIV-1 in a Latent State via Chromatin-mediated Transcriptional Silencing
- PMID: 26506039
- PMCID: PMC4881759
- DOI: 10.1038/mtna.2015.31
Novel RNA Duplex Locks HIV-1 in a Latent State via Chromatin-mediated Transcriptional Silencing
Abstract
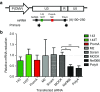
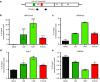
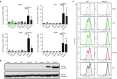
Transcriptional gene silencing (TGS) of mammalian genes can be induced by short interfering RNA (siRNA) targeting promoter regions. We previously reported potent TGS of HIV-1 by siRNA (PromA), which targets tandem NF-κB motifs within the viral 5'LTR. In this study, we screened a siRNA panel with the aim of identifying novel 5'LTR targets, to provide multiplexing potential with enhanced viral silencing and application toward developing alternate therapeutic strategies. Systematic examination identified a novel siRNA target, si143, confirmed to induce TGS as the silencing mechanism. TGS was prolonged with virus suppression >12 days, despite a limited ability to induce post- TGS. Epigenetic changes associated with silencing were suggested by partial reversal by histone deacetylase inhibitors and confirmed by chromatin immunoprecipitation analyses, which showed induction of H3K27me3 and H3K9me3, reduction in H3K9Ac, and recruitment of argonaute-1, all characteristic marks of heterochromatin and TGS. Together, these epigenetic changes mimic those associated with HIV-1 latency. Further, robust resistance to reactivation was observed in the J-Lat 9.2 cell latency model, when transduced with shPromA and/or sh143. These data support si/shRNA-mediated TGS approaches to HIV-1 and provide alternate targets to pursue a functional cure, whereby the viral reservoir is locked in latency following antiretroviral therapy cessation.
Figures






References
-
- Wong, JK, Hezareh, M, Günthard, HF, Havlir, DV, Ignacio, CC, Spina, CA et al. (1997). Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278: 1291–1295. - PubMed
-
- Gandhi, RT, Bosch, RJ, Aga, E, Albrecht, M, Demeter, LM, Dykes, C et al.; AIDS Clinical Trials Group A5173 Team. (2010). No evidence for decay of the latent reservoir in HIV-1-infected patients receiving intensive enfuvirtide-containing antiretroviral therapy. J Infect Dis 201: 293–296. - PMC - PubMed
-
- Gandhi, RT, Zheng, L, Bosch, RJ, Chan, ES, Margolis, DM, Read, S et al.; AIDS Clinical Trials Group A5244 Team. (2010). The effect of raltegravir intensification on low-level residual viremia in HIV-infected patients on antiretroviral therapy: a randomized controlled trial. PLoS Med 7. - PMC - PubMed
-
- Hammer, SM, Ribaudo, H, Bassett, R, Mellors, JW, Demeter, LM, Coombs, RW et al.; AIDS Clinical Trials Group (ACTG) 372A Study Team. (2010). A randomized, placebo-controlled trial of abacavir intensification in HIV-1-infected adults with virologic suppression on a protease inhibitor-containing regimen. HIV Clin Trials 11: 312–324. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

