Origins of Programmable Nucleases for Genome Engineering
- PMID: 26506267
- PMCID: PMC4798875
- DOI: 10.1016/j.jmb.2015.10.014
Origins of Programmable Nucleases for Genome Engineering
Abstract
Genome engineering with programmable nucleases depends on cellular responses to a targeted double-strand break (DSB). The first truly targetable reagents were the zinc finger nucleases (ZFNs) showing that arbitrary DNA sequences could be addressed for cleavage by protein engineering, ushering in the breakthrough in genome manipulation. ZFNs resulted from basic research on zinc finger proteins and the FokI restriction enzyme (which revealed a bipartite structure with a separable DNA-binding domain and a non-specific cleavage domain). Studies on the mechanism of cleavage by 3-finger ZFNs established that the preferred substrates were paired binding sites, which doubled the size of the target sequence recognition from 9 to 18bp, long enough to specify a unique genomic locus in plant and mammalian cells. Soon afterwards, a ZFN-induced DSB was shown to stimulate homologous recombination in cells. Transcription activator-like effector nucleases (TALENs) that are based on bacterial TALEs fused to the FokI cleavage domain expanded this capability. The fact that ZFNs and TALENs have been used for genome modification of more than 40 different organisms and cell types attests to the success of protein engineering. The most recent technology platform for delivering a targeted DSB to cellular genomes is that of the RNA-guided nucleases, which are based on the naturally occurring Type II prokaryotic CRISPR-Cas9 system. Unlike ZFNs and TALENs that use protein motifs for DNA sequence recognition, CRISPR-Cas9 depends on RNA-DNA recognition. The advantages of the CRISPR-Cas9 system-the ease of RNA design for new targets and the dependence on a single, constant Cas9 protein-have led to its wide adoption by research laboratories around the world. These technology platforms have equipped scientists with an unprecedented ability to modify cells and organisms almost at will, with wide-ranging implications across biology and medicine. However, these nucleases have also been shown to cut at off-target sites with mutagenic consequences. Therefore, issues such as efficacy, specificity and delivery are likely to drive selection of reagents for particular purposes. Human therapeutic applications of these technologies will ultimately depend on risk versus benefit analysis and informed consent.
Keywords: CRISPR-Cas9; gene therapy; genome editing; transcription activator-like effector nucleases (TALENs); zinc finger nucleases (ZFNs).
Copyright © 2015 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures


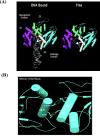
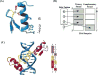




Comment in
-
What history tells us XLIV: The construction of the zinc finger nucleases.J Biosci. 2017 Dec;42(4):527-530. doi: 10.1007/s12038-017-9723-4. J Biosci. 2017. PMID: 29229870 No abstract available.
References
-
- Mansour SL, Thomas KR, Capecchi MR. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature. 1988;336(6197):348–352. - PubMed
-
- Capecchi MR. Altering the genome by homologous recombination. Science. 1989;244:1288–1292. - PubMed
-
- Kita K, Kotani H, Sugisaki H, Takanami M. The FokI restriction-modification systems. I Organization and nucleotide sequences of the restriction and modification genes. J Biol Chem. 1989;264:5751–5756. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

