Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species
- PMID: 26506948
- PMCID: PMC4623524
- DOI: 10.1038/srep15655
Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species
Abstract
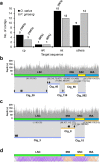
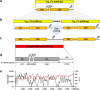
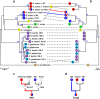
Cytoplasmic chloroplast (cp) genomes and nuclear ribosomal DNA (nR) are the primary sequences used to understand plant diversity and evolution. We introduce a high-throughput method to simultaneously obtain complete cp and nR sequences using Illumina platform whole-genome sequence. We applied the method to 30 rice specimens belonging to nine Oryza species. Concurrent phylogenomic analysis using cp and nR of several of specimens of the same Oryza AA genome species provides insight into the evolution and domestication of cultivated rice, clarifying three ambiguous but important issues in the evolution of wild Oryza species. First, cp-based trees clearly classify each lineage but can be biased by inter-subspecies cross-hybridization events during speciation. Second, O. glumaepatula, a South American wild rice, includes two cytoplasm types, one of which is derived from a recent interspecies hybridization with O. longistminata. Third, the Australian O. rufipogan-type rice is a perennial form of O. meridionalis.
Figures

References
-
- Qiu Y.-L. et al. The earliest angiosperms: evidence from mitochondrial, plastid and nuclear genomes. Nature 402, 404–407 (1999). - PubMed
-
- Soltis P. S., Soltis D. E. & Chase M. W. Angiosperm phylogeny inferred from multiple genes as a tool for comparative biology. Nature 402, 402–404 (1999). - PubMed
-
- Park J. Y. et al. Complete mitochondrial genome sequence and identification of a candidate gene responsible for cytoplasmic male sterility in radish (Raphanus sativus L.) containing DCGMS cytoplasm. Theor. Appl. Genet. 126, 1763–1774 (2013). - PubMed
-
- Palmer J. D. Comparative organization of chloroplast genomes. Annu. Rev. Genet. 19, 325–354 (1985). - PubMed
-
- Harris S. A. & Ingram R. Chloroplast DNA and biosystematics: The effects of intraspecific diversity and plastid transmission. Taxon 40, 393–412 (1991).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

