DNA N(6)-methyladenine: a new epigenetic mark in eukaryotes?
- PMID: 26507168
- PMCID: PMC4763336
- DOI: 10.1038/nrm4076
DNA N(6)-methyladenine: a new epigenetic mark in eukaryotes?
Abstract
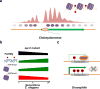
DNA N(6)-adenine methylation (N(6)-methyladenine; 6mA) in prokaryotes functions primarily in the host defence system. The prevalence and significance of this modification in eukaryotes had been unclear until recently. Here, we discuss recent publications documenting the presence of 6mA in Chlamydomonas reinhardtii, Drosophila melanogaster and Caenorhabditis elegans; consider possible roles for this DNA modification in regulating transcription, the activity of transposable elements and transgenerational epigenetic inheritance; and propose 6mA as a new epigenetic mark in eukaryotes.
Conflict of interest statement
YS is a co-founder of Constellation Pharmaceuticals, Inc and a member of its scientific advisory board. The other authors declare no competing interests.
Figures
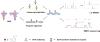

References
-
- Vanyushin BF, Tkacheva SG, Belozersky AN. Rare bases in animal DNA. Nature. 1970;225:948–9. - PubMed
-
- Dunn DB, Smith JD. Occurrence of a new base in the deoxyribonucleic acid of a strain of Bacterium coli. Nature. 1955;175:336–7. - PubMed
-
- Vanyushin BF, Belozersky AN, Kokurina NA, Kadirova DX. 5-methylcytosine and 6-methylamino-purine in bacterial DNA. Nature. 1968;218:1066–7. - PubMed
-
- Unger G, Venner H. Remarks on minor bases in spermatic desoxyribonucleic acid. Hoppe Seylers Z Physiol Chem. 1966;344:280–3. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

