BloodSpot: a database of gene expression profiles and transcriptional programs for healthy and malignant haematopoiesis
- PMID: 26507857
- PMCID: PMC4702803
- DOI: 10.1093/nar/gkv1101
BloodSpot: a database of gene expression profiles and transcriptional programs for healthy and malignant haematopoiesis
Abstract
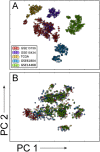
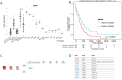
Research on human and murine haematopoiesis has resulted in a vast number of gene-expression data sets that can potentially answer questions regarding normal and aberrant blood formation. To researchers and clinicians with limited bioinformatics experience, these data have remained available, yet largely inaccessible. Current databases provide information about gene-expression but fail to answer key questions regarding co-regulation, genetic programs or effect on patient survival. To address these shortcomings, we present BloodSpot (www.bloodspot.eu), which includes and greatly extends our previously released database HemaExplorer, a database of gene expression profiles from FACS sorted healthy and malignant haematopoietic cells. A revised interactive interface simultaneously provides a plot of gene expression along with a Kaplan-Meier analysis and a hierarchical tree depicting the relationship between different cell types in the database. The database now includes 23 high-quality curated data sets relevant to normal and malignant blood formation and, in addition, we have assembled and built a unique integrated data set, BloodPool. Bloodpool contains more than 2000 samples assembled from six independent studies on acute myeloid leukemia. Furthermore, we have devised a robust sample integration procedure that allows for sensitive comparison of user-supplied patient samples in a well-defined haematopoietic cellular space.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

