Ubiquitin-Activated Interaction Traps (UBAITs) identify E3 ligase binding partners
- PMID: 26508657
- PMCID: PMC4693525
- DOI: 10.15252/embr.201540620
Ubiquitin-Activated Interaction Traps (UBAITs) identify E3 ligase binding partners
Abstract
We describe a new class of reagents for identifying substrates, adaptors, and regulators of HECT and RING E3s. UBAITs (Ubiquitin-Activated Interaction Traps) are E3-ubiquitin fusion proteins and, in an E1- and E2-dependent manner, the C-terminal ubiquitin moiety forms an amide linkage to proteins that interact with the E3, enabling covalent co-purification of the E3 with partner proteins. We designed UBAITs for both HECT (Rsp5, Itch) and RING (Psh1, RNF126, RNF168) E3s. For HECT E3s, trapping of interacting proteins occurred in vitro either through an E3 thioester-linked lariat intermediate or through an E2 thioester intermediate, and both WT and active-site mutant UBAITs trapped known interacting proteins in yeast and human cells. Yeast Psh1 and human RNF126 and RNF168 UBAITs also trapped known interacting proteins when expressed in cells. Human RNF168 is a key mediator of ubiquitin signaling that promotes DNA double-strand break repair. Using the RNF168 UBAIT, we identify H2AZ--a histone protein involved in DNA repair--as a new target of this E3 ligase. These results demonstrate that UBAITs represent powerful tools for profiling a wide range of ubiquitin ligases.
Keywords: HECT E3s; RING E3s; Ubiquitin; Ubiquitin ligases.
© 2015 The Authors.
Figures
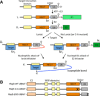
Schematic of proposed UBAIT mechanism. The E1 enzyme activates the UBAIT, forming a thioester‐linked complex (i.), which is then transferred to an E2 enzyme (ii.). The active‐site cysteine of the E3 can attack the E2 thioester, forming a thioester‐linked protein lariat structure (iii.); when an amine group of a target protein attacks the thioester bond, a stable amide linkage is formed between the UBAIT and the target protein (v.). If the UBAIT does not form a lariat structure, as in the case of the active‐site mutant (iv.), an amine group of a target protein can potentially directly attack the E2 thioester bond, also resulting in a covalent UBAIT‐target protein complex (v.).
Depiction of the wild‐type Rsp5 UBAIT (WT), active‐site cysteine mutant (C‐A) UBAIT, and ΔGG ubiquitin (ΔGG) UBAIT.
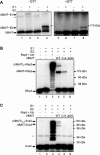
Rsp5 can be activated by E1 and transferred to an E2. Purified 32P‐labeled Rsp5 UBAIT (WT) was incubated with or without E1 and E2 enzymes, and products were analyzed by SDS–PAGE in the absence or presence of DTT in the loading buffer. The migration points of the UBAIT˜E1 and UBAIT˜E2 thioesters are indicated. The asterisk marks a contaminant of the purification process (uncut GST‐tagged UBAIT).
Rsp5 UBAITs were incubated with E1 and E2 proteins, as indicated, and 32P‐labeled Wbp2. The migration points of the major products are indicated with arrows.
Rsp5 UBAITs were incubated with E1 and E2 proteins, as indicated, and 32P‐labeled Sna3. The migration points of the major products are indicated with arrows. The asterisk marks a contaminant of the purification process (uncut GST‐Sna3).

Depiction of wild‐type RNF168, RNF168 UBAIT (WT), and ΔGG ubiquitin (ΔGG) UBAIT. The structural positions of the ubiquitin binding domains (UBDs) MIU (motif interacting with ubiquitin), UMI (UIM‐ and MIU‐related UBD), and RING domains are indicated.
Immunoblot (anti‐TAP) of extracts from human HEK293T cells expressing TAP‐RNF168 or the indicated TAP‐RNF168 UBAITs.
Ubiquitylation of FLAG‐H2AZ in HEK293T cells transiently transfected with FLAG‐H2AZ in the presence and absence of MYC‐RNF168 (lanes 1 and 2, respectively; lower panel). Expression of MYC‐RNF168 was verified by anti‐MYC immunoblotting (upper panel).
Purified H2AZ‐containing reconstituted nucleosome core particles (NCPs) were used in an in vitro ubiquitylation assay in the presence of RNF168. WT H2AZ‐containing NCPs (first two lanes) were incubated with RNF168, UBCH5a, and ubiquitin, without or with E1 enzyme (lanes 1 and 2, respectively). Third lane contains D94A‐containing NCPs, incubated with RNF168, UbcH5a, ubiquitin, and E1.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

