CLL Exosomes Modulate the Transcriptome and Behaviour of Recipient Stromal Cells and Are Selectively Enriched in miR-202-3p
- PMID: 26509439
- PMCID: PMC4625016
- DOI: 10.1371/journal.pone.0141429
CLL Exosomes Modulate the Transcriptome and Behaviour of Recipient Stromal Cells and Are Selectively Enriched in miR-202-3p
Abstract
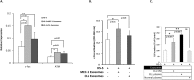
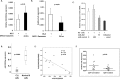
Bi-directional communication with the microenvironment is essential for homing and survival of cancer cells with implications for disease biology and behaviour. In chronic lymphocytic leukemia (CLL), the role of the microenvironment on malignant cell behaviour is well described. However, how CLL cells engage and recruit nurturing cells is poorly characterised. Here we demonstrate that CLL cells secrete exosomes that are nanovesicles originating from the fusion of multivesicular bodies with the plasma membrane, to shuttle proteins, lipids, microRNAs (miR) and mRNAs to recipient cells. We characterise and confirm the size (50-100 nm) and identity of the CLL-derived exosomes by Electron microscopy (EM), Atomic force microscopy (AFM), flow cytometry and western blotting using both exosome- and CLL-specific markers. Incubation of CLL-exosomes, derived either from cell culture supernatants or from patient plasma, with human stromal cells shows that they are readily taken up into endosomes, and induce expression of genes such as c-fos and ATM as well as enhance proliferation of recipient HS-5 cells. Furthermore, we show that CLL exosomes encapsulate abundant small RNAs and are enriched in certain miRs and specifically hsa-miR-202-3p. We suggest that such specific packaging of miR-202-3p into exosomes results in enhanced expression of 'suppressor of fused' (Sufu), a Hedgehog (Hh) signalling intermediate, in the parental CLL cells. Thus, our data show that CLL cells secrete exosomes that alter the transcriptome and behaviour of recipient cells. Such communication with microenvironment is likely to have an important role in CLL disease biology.
Conflict of interest statement
Figures




Similar articles
-
Characterization of CLL exosomes reveals a distinct microRNA signature and enhanced secretion by activation of BCR signaling.Blood. 2015 May 21;125(21):3297-305. doi: 10.1182/blood-2014-12-618470. Epub 2015 Apr 1. Blood. 2015. PMID: 25833959 Free PMC article.
-
Exosomes derived from chronic lymphocytic leukaemia cells transfer miR-146a to induce the transition of mesenchymal stromal cells into cancer-associated fibroblasts.J Biochem. 2020 Nov 1;168(5):491-498. doi: 10.1093/jb/mvaa064. J Biochem. 2020. PMID: 32770182
-
IL-4 Up-Regulates MiR-21 and the MiRNAs Hosted in the CLCN5 Gene in Chronic Lymphocytic Leukemia.PLoS One. 2015 Apr 24;10(4):e0124936. doi: 10.1371/journal.pone.0124936. eCollection 2015. PLoS One. 2015. PMID: 25909590 Free PMC article.
-
Exosome-derived microRNAs in cancer metabolism: possible implications in cancer diagnostics and therapy.Exp Mol Med. 2017 Jan 20;49(1):e285. doi: 10.1038/emm.2016.153. Exp Mol Med. 2017. PMID: 28104913 Free PMC article. Review.
-
Exosome-carried microRNA-based signature as a cellular trigger for the evolution of chronic lymphocytic leukemia into Richter syndrome.Crit Rev Clin Lab Sci. 2018 Nov;55(7):501-515. doi: 10.1080/10408363.2018.1499707. Epub 2018 Sep 21. Crit Rev Clin Lab Sci. 2018. PMID: 30238808 Review.
Cited by
-
Circulating miRNAs in Breast Cancer Diagnosis and Prognosis.Cancers (Basel). 2022 May 7;14(9):2317. doi: 10.3390/cancers14092317. Cancers (Basel). 2022. PMID: 35565446 Free PMC article. Review.
-
The emerging roles of tumor-derived exosomes in hematological malignancies.Leukemia. 2017 Jun;31(6):1259-1268. doi: 10.1038/leu.2017.91. Epub 2017 Mar 21. Leukemia. 2017. PMID: 28321122 Review.
-
The emerging roles of exosomes in tumor-stroma interaction.J Cancer Res Clin Oncol. 2016 Sep;142(9):1897-907. doi: 10.1007/s00432-016-2145-0. Epub 2016 Mar 17. J Cancer Res Clin Oncol. 2016. PMID: 26987524 Free PMC article. Review.
-
Ultracentrifugation versus kit exosome isolation: nanoLC-MS and other tools reveal similar performance biomarkers, but also contaminations.Future Sci OA. 2018 Nov 9;5(1):FSO359. doi: 10.4155/fsoa-2018-0088. eCollection 2019 Jan. Future Sci OA. 2018. PMID: 30652024 Free PMC article.
-
The Landscape of Exosome-Derived Non-Coding RNA in Leukemia.Front Pharmacol. 2022 Jun 15;13:912303. doi: 10.3389/fphar.2022.912303. eCollection 2022. Front Pharmacol. 2022. PMID: 35784717 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

