Structural basis for phosphatidylinositol-phosphate biosynthesis
- PMID: 26510127
- PMCID: PMC4634129
- DOI: 10.1038/ncomms9505
Structural basis for phosphatidylinositol-phosphate biosynthesis
Abstract
Phosphatidylinositol is critical for intracellular signalling and anchoring of carbohydrates and proteins to outer cellular membranes. The defining step in phosphatidylinositol biosynthesis is catalysed by CDP-alcohol phosphotransferases, transmembrane enzymes that use CDP-diacylglycerol as donor substrate for this reaction, and either inositol in eukaryotes or inositol phosphate in prokaryotes as the acceptor alcohol. Here we report the structures of a related enzyme, the phosphatidylinositol-phosphate synthase from Renibacterium salmoninarum, with and without bound CDP-diacylglycerol to 3.6 and 2.5 Å resolution, respectively. These structures reveal the location of the acceptor site, and the molecular determinants of substrate specificity and catalysis. Functional characterization of the 40%-identical ortholog from Mycobacterium tuberculosis, a potential target for the development of novel anti-tuberculosis drugs, supports the proposed mechanism of substrate binding and catalysis. This work therefore provides a structural and functional framework to understand the mechanism of phosphatidylinositol-phosphate biosynthesis.
Figures


 ions labelled and depicted in stick representation.
ions labelled and depicted in stick representation.
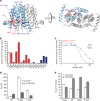
 (closed diamonds) and
(closed diamonds) and  (open circles) inhibit the activity of MtPIPS-FL with half-inhibitory concentrations of 44 and 22 mM, respectively. (d) KM of MtPIPS-FL WT, R195Q and P153V for inositol phosphate (InsP; white) and CDP-DAG (grey). InsP,
(open circles) inhibit the activity of MtPIPS-FL with half-inhibitory concentrations of 44 and 22 mM, respectively. (d) KM of MtPIPS-FL WT, R195Q and P153V for inositol phosphate (InsP; white) and CDP-DAG (grey). InsP, References
-
- Umesiri F. E., Sanki A. K., Boucau J., Ronning D. R. & Sucheck S. J. Recent advances toward the inhibition of mAG and LAM synthesis in Mycobacterium tuberculosis. Med. Res. Rev. 30, 290–326 (2010). - PubMed
-
- Morii H., Ogawa M., Fukuda K., Taniguchi H. & Koga Y. A revised biosynthetic pathway for phosphatidylinositol in Mycobacteria. J. Biochem. 148, 593–602 (2010). - PubMed
-
- Morii H., Kiyonari S., Ishino Y. & Koga Y. A novel biosynthetic pathway of archaetidyl-myo-inositol via archaetidyl-myo-inositol phosphate from CDP-archaeol and D-glucose 6-phosphate in methanoarchaeon Methanothermobacter thermautotrophicus cells. J. Biol. Chem. 284, 30766–30774 (2009). - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

