Basal autophagy maintains pancreatic acinar cell homeostasis and protein synthesis and prevents ER stress
- PMID: 26512112
- PMCID: PMC4653219
- DOI: 10.1073/pnas.1519384112
Basal autophagy maintains pancreatic acinar cell homeostasis and protein synthesis and prevents ER stress
Abstract
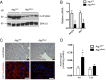
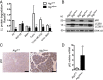
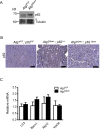
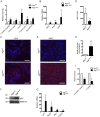
Pancreatic acinar cells possess very high protein synthetic rates as they need to produce and secrete large amounts of digestive enzymes. Acinar cell damage and dysfunction cause malnutrition and pancreatitis, and inflammation of the exocrine pancreas that promotes development of pancreatic ductal adenocarcinoma (PDAC), a deadly pancreatic neoplasm. The cellular and molecular mechanisms that maintain acinar cell function and whose dysregulation can lead to tissue damage and chronic pancreatitis are poorly understood. It was suggested that autophagy, the principal cellular degradative pathway, is impaired in pancreatitis, but it is unknown whether impaired autophagy is a cause or a consequence of pancreatitis. To address this question, we generated Atg7(Δpan) mice that lack the essential autophagy-related protein 7 (ATG7) in pancreatic epithelial cells. Atg7(Δpan) mice exhibit severe acinar cell degeneration, leading to pancreatic inflammation and extensive fibrosis. Whereas ATG7 loss leads to the expected decrease in autophagic flux, it also results in endoplasmic reticulum (ER) stress, accumulation of dysfunctional mitochondria, oxidative stress, activation of AMPK, and a marked decrease in protein synthetic capacity that is accompanied by loss of rough ER. Atg7(Δpan) mice also exhibit spontaneous activation of regenerative mechanisms that initiate acinar-to-ductal metaplasia (ADM), a process that replaces damaged acinar cells with duct-like structures.
Keywords: ATG7; autophagy; pancreatitis; protein synthesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




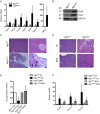
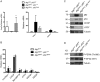
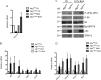
References
-
- Case RM. Synthesis, intracellular transport and discharge of exportable proteins in the pancreatic acinar cell and other cells. Biol Rev Camb Philos Soc. 1978;53(2):211–354. - PubMed
-
- Wang M, Kaufman RJ. The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat Rev Cancer. 2014;14(9):581–597. - PubMed
-
- LaRusch J, Solomon S, Whitcomb DC. 1993. Pancreatitis Overview, eds Pagon RA, et al. (GeneReviews, University of Washington, Seattle)
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

