The standard operating procedure of the DOE-JGI Microbial Genome Annotation Pipeline (MGAP v.4)
- PMID: 26512311
- PMCID: PMC4623924
- DOI: 10.1186/s40793-015-0077-y
The standard operating procedure of the DOE-JGI Microbial Genome Annotation Pipeline (MGAP v.4)
Erratum in
-
Erratum to: The standard operating procedure of the DOE-JGI Microbial Genome Annotation Pipeline (MGAP v.4).Stand Genomic Sci. 2016 Mar 21;11:27. doi: 10.1186/s40793-016-0148-8. eCollection 2016. Stand Genomic Sci. 2016. PMID: 27004084 Free PMC article.
Abstract
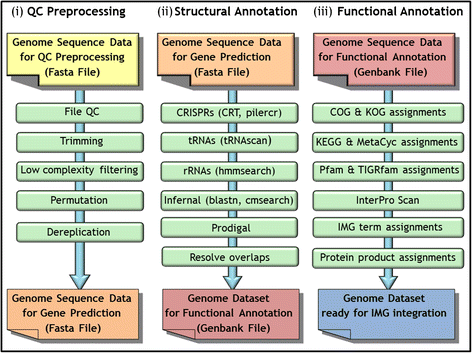
The DOE-JGI Microbial Genome Annotation Pipeline performs structural and functional annotation of microbial genomes that are further included into the Integrated Microbial Genome comparative analysis system. MGAP is applied to assembled nucleotide sequence datasets that are provided via the IMG submission site. Dataset submission for annotation first requires project and associated metadata description in GOLD. The MGAP sequence data processing consists of feature prediction including identification of protein-coding genes, non-coding RNAs and regulatory RNA features, as well as CRISPR elements. Structural annotation is followed by assignment of protein product names and functions.
Keywords: IMG; JGI; Microbial Genome Annotation; SOP.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources