Deficient and Null Variants of SERPINA1 Are Proteotoxic in a Caenorhabditis elegans Model of α1-Antitrypsin Deficiency
- PMID: 26512890
- PMCID: PMC4626213
- DOI: 10.1371/journal.pone.0141542
Deficient and Null Variants of SERPINA1 Are Proteotoxic in a Caenorhabditis elegans Model of α1-Antitrypsin Deficiency
Abstract
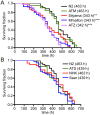
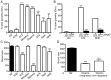
α1-antitrypsin deficiency (ATD) predisposes patients to both loss-of-function (emphysema) and gain-of-function (liver cirrhosis) phenotypes depending on the type of mutation. Although the Z mutation (ATZ) is the most prevalent cause of ATD, >120 mutant alleles have been identified. In general, these mutations are classified as deficient (<20% normal plasma levels) or null (<1% normal levels) alleles. The deficient alleles, like ATZ, misfold in the ER where they accumulate as toxic monomers, oligomers and aggregates. Thus, deficient alleles may predispose to both gain- and loss-of-function phenotypes. Null variants, if translated, typically yield truncated proteins that are efficiently degraded after being transiently retained in the ER. Clinically, null alleles are only associated with the loss-of-function phenotype. We recently developed a C. elegans model of ATD in order to further elucidate the mechanisms of proteotoxicity (gain-of-function phenotype) induced by the aggregation-prone deficient allele, ATZ. The goal of this study was to use this C. elegans model to determine whether different types of deficient and null alleles, which differentially affect polymerization and secretion rates, correlated to any extent with proteotoxicity. Animals expressing the deficient alleles, Mmalton, Siiyama and S (ATS), showed overall toxicity comparable to that observed in patients. Interestingly, Siiyama expressing animals had smaller intracellular inclusions than ATZ yet appeared to have a greater negative effect on animal fitness. Surprisingly, the null mutants, although efficiently degraded, showed a relatively mild gain-of-function proteotoxic phenotype. However, since null variant proteins are degraded differently and do not appear to accumulate, their mechanism of proteotoxicity is likely to be different to that of polymerizing, deficient mutants. Taken together, these studies showed that C. elegans is an inexpensive tool to assess the proteotoxicity of different AT variants using a transgenic approach.
Conflict of interest statement
Figures
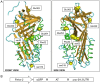
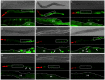

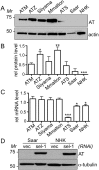
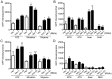
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

