Combining Search, Social Media, and Traditional Data Sources to Improve Influenza Surveillance
- PMID: 26513245
- PMCID: PMC4626021
- DOI: 10.1371/journal.pcbi.1004513
Combining Search, Social Media, and Traditional Data Sources to Improve Influenza Surveillance
Abstract
We present a machine learning-based methodology capable of providing real-time ("nowcast") and forecast estimates of influenza activity in the US by leveraging data from multiple data sources including: Google searches, Twitter microblogs, nearly real-time hospital visit records, and data from a participatory surveillance system. Our main contribution consists of combining multiple influenza-like illnesses (ILI) activity estimates, generated independently with each data source, into a single prediction of ILI utilizing machine learning ensemble approaches. Our methodology exploits the information in each data source and produces accurate weekly ILI predictions for up to four weeks ahead of the release of CDC's ILI reports. We evaluate the predictive ability of our ensemble approach during the 2013-2014 (retrospective) and 2014-2015 (live) flu seasons for each of the four weekly time horizons. Our ensemble approach demonstrates several advantages: (1) our ensemble method's predictions outperform every prediction using each data source independently, (2) our methodology can produce predictions one week ahead of GFT's real-time estimates with comparable accuracy, and (3) our two and three week forecast estimates have comparable accuracy to real-time predictions using an autoregressive model. Moreover, our results show that considerable insight is gained from incorporating disparate data streams, in the form of social media and crowd sourced data, into influenza predictions in all time horizons.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
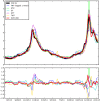
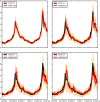

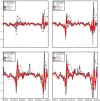
References
-
- WHO (2015) Influenza (Seasonal), Fact Sheet Number 211. Available at http://www.who.int/mediacentre/factsheets/fs211/en/index.html.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

