Robust and efficient parameter estimation in dynamic models of biological systems
- PMID: 26515482
- PMCID: PMC4625902
- DOI: 10.1186/s12918-015-0219-2
Robust and efficient parameter estimation in dynamic models of biological systems
Abstract
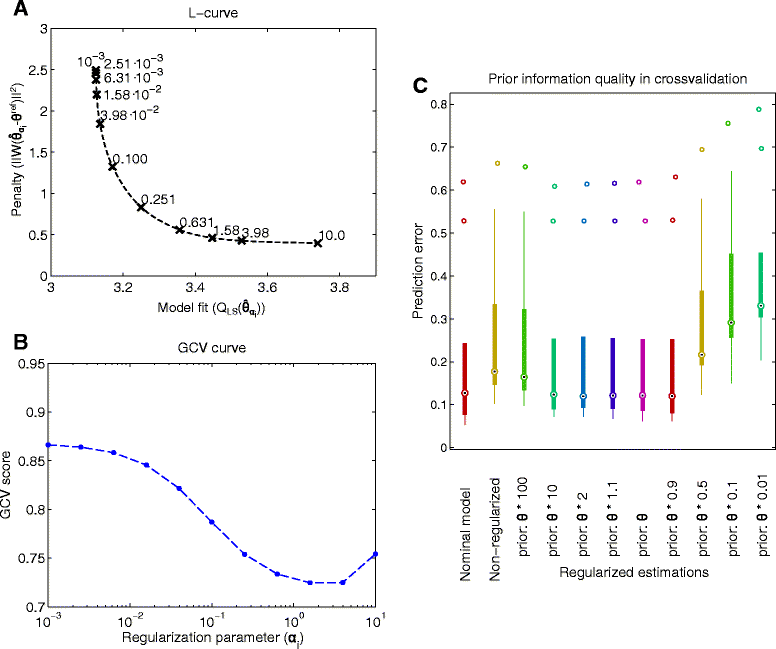
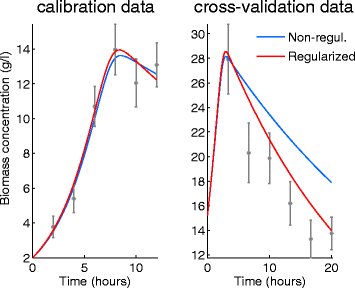
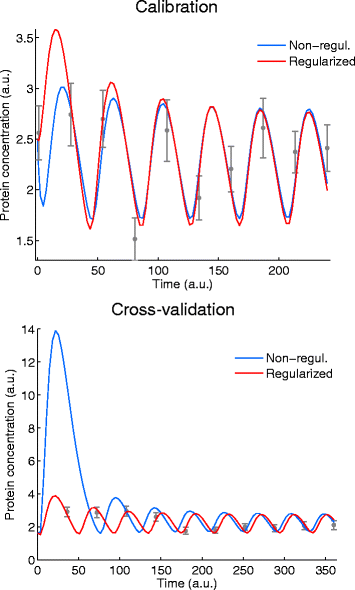
Background: Dynamic modelling provides a systematic framework to understand function in biological systems. Parameter estimation in nonlinear dynamic models remains a very challenging inverse problem due to its nonconvexity and ill-conditioning. Associated issues like overfitting and local solutions are usually not properly addressed in the systems biology literature despite their importance. Here we present a method for robust and efficient parameter estimation which uses two main strategies to surmount the aforementioned difficulties: (i) efficient global optimization to deal with nonconvexity, and (ii) proper regularization methods to handle ill-conditioning. In the case of regularization, we present a detailed critical comparison of methods and guidelines for properly tuning them. Further, we show how regularized estimations ensure the best trade-offs between bias and variance, reducing overfitting, and allowing the incorporation of prior knowledge in a systematic way.
Results: We illustrate the performance of the presented method with seven case studies of different nature and increasing complexity, considering several scenarios of data availability, measurement noise and prior knowledge. We show how our method ensures improved estimations with faster and more stable convergence. We also show how the calibrated models are more generalizable. Finally, we give a set of simple guidelines to apply this strategy to a wide variety of calibration problems.
Conclusions: Here we provide a parameter estimation strategy which combines efficient global optimization with a regularization scheme. This method is able to calibrate dynamic models in an efficient and robust way, effectively fighting overfitting and allowing the incorporation of prior information.
Figures
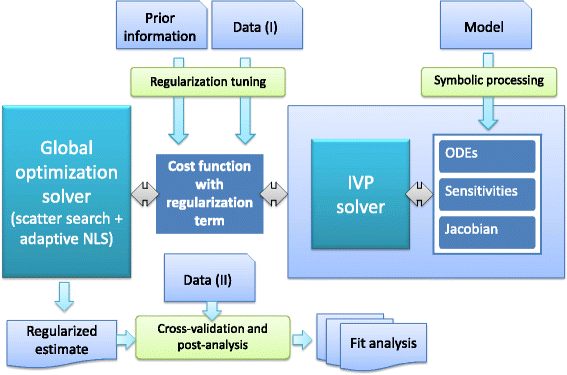
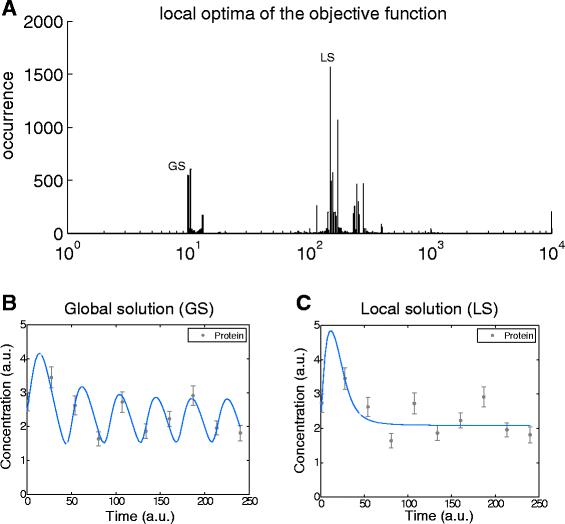
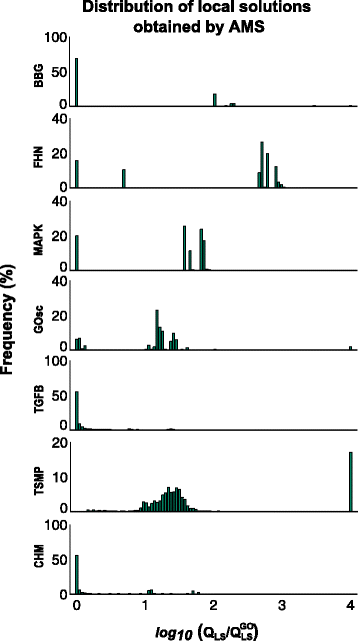
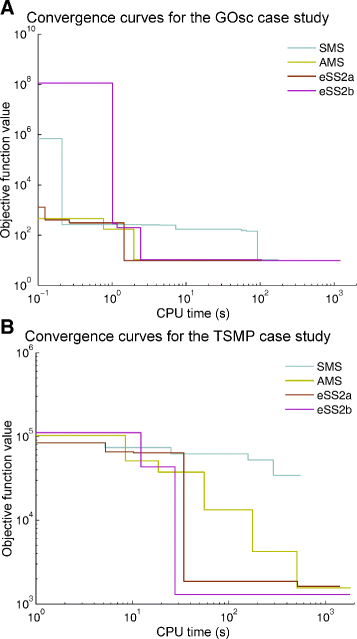



References
-
- Epstein JM. Why model? J Artif Soc Social Simul. 2008;11(4):12.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

