JuncDB: an exon-exon junction database
- PMID: 26519469
- PMCID: PMC4702826
- DOI: 10.1093/nar/gkv1142
JuncDB: an exon-exon junction database
Abstract
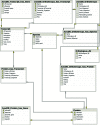
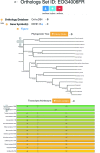
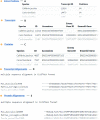
Intron positions upon the mRNA transcript are sometimes remarkably conserved even across distantly related eukaryotic species. This has made the comparison of intron-exon architectures across orthologous transcripts a very useful tool for studying various evolutionary processes. Moreover, the wide range of functions associated with introns may confer biological meaning to evolutionary changes in gene architectures. Yet, there is currently no database that offers such comparative information. Here, we present JuncDB (http://juncdb.carmelab.huji.ac.il/), an exon-exon junction database dedicated to the comparison of architectures between orthologous transcripts. It covers nearly 40,000 sets of orthologous transcripts spanning 88 eukaryotic species. JuncDB offers a user-friendly interface, access to detailed information, instructive graphical displays of the comparative data and easy ways to download data to a local computer. In addition, JuncDB allows the analysis to be carried out either on specific genes, or at a genome-wide level for any selected group of species.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Rogozin I.B., Wolf Y.I., Sorokin A.V., Mirkin B.G., Koonin E.V. Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr. Biol. 2003;13:1512–1517. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

